Found 768 results
Open Access
Review
09 September 2024Application of Polydopamine-Based Photocatalysts in Energy and Environmental Systems
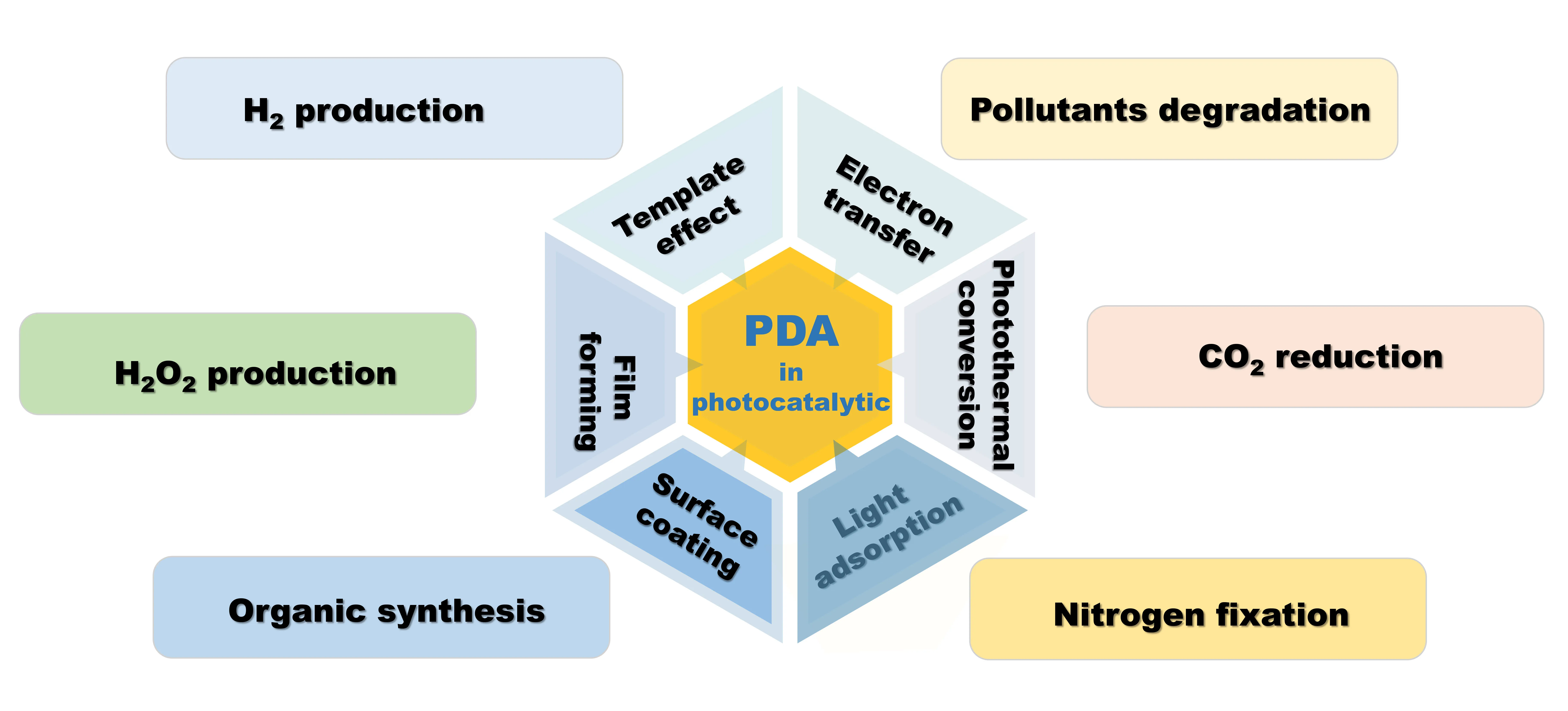
Polydopamine (PDA) is also widely sought after in photocatalytic applications due to its fascinating properties such as simple preparation, templating agent, near-infrared absorption, high photothermal conversion efficiency, abundant functional groups, and strong chelating effect of metal ions. This review will present the structural features and synthetic methods of PDA, the advantages of PDA for photocatalytic applications (templating agent effect, light absorption properties, film-forming properties, hydrophilicity, conductivity, etc.), the modulation strategies of PDA for photocatalytic applications, and the use of PDA-based photocatalytic materials for solar-powered water purification (heavy metal adsorption and reduction, catalytic degradation of organic pollutants, and antimicrobial properties), hydrogen production, hydrogen peroxide production, CO2 reduction, and organic conversion. Finally, this review will provide valuable information for the design and development of PDA-based photocatalytic materials.
Open Access
Perspective
09 September 2024Social, Ecological and Economic Synergies of Forests for Sustainability Contradict Projects Involving Large-Scale Deforestation for Energy Production
Good projects and solutions aiming at sustainable development must repair the damage done in past decades by being explicitly designed and monitored to achieve synergetic benefits for the environment and society. We identify environmental, social and economic aspects of sustainability in which enlightened forest management can increase the fulfillment of human and ecological needs and hence the quality of life of present and future generations. Projects aiming at energy production and profits at the cost of biodiversity, nature protection, and human health and well-being are therefore questionable and increasingly socially and politically unacceptable—especially where the viability of alternative options with better social and ecological footprints can be easily demonstrated. This is also true for renewable energy projects. The perspective presented here demonstrates how ostensibly renewable energy projects in natural areas, such as large-scale wind and solar power plants in traditional forests, which are planned, for example, in Germany, may be detrimental to ecological and social sustainability. Forests cut down for such projects are “non-renewable” within reasonable time-scales left to stabilize our climate and ecosystems. Such projects also impair the credibility of the proclaimed role model character and sustainability leadership of Global North countries, which can lead to negative implications for the protection of forests in tropical countries.
Open Access
Review
06 September 2024Principles in Identification of Human Remains through Forensic Odontology
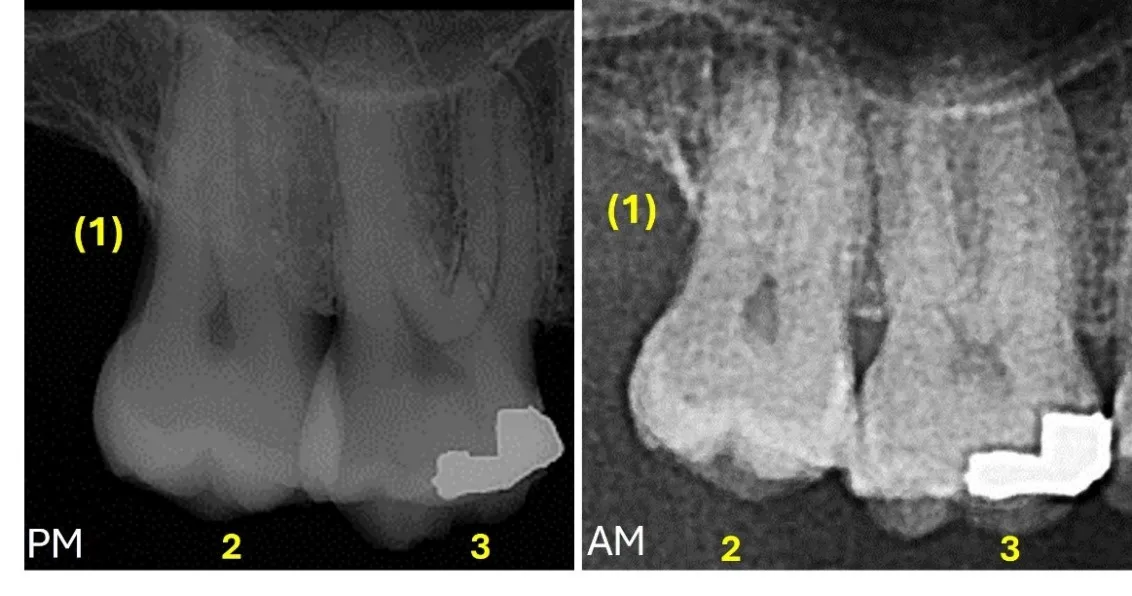
Human identification is the core component of Forensic Odontology. The process of identification of unknown remains generally starts with the reconstruction of the skeletal biological profile, which provides a general description of the individual that is used to narrow down the candidates for the identity. Once one or more candidates are given for the identity, forensic odontologists conduct the comparison between the antemortem and postmortem records. The postmortem vs. the antemortem data comparison implies the evaluation of the consistencies and inconsistencies found in the data sets. This comparison is highly affected by the quality and completeness of the antemortem records, as well as the condition of the remains. The principles of the odontological comparison are based on the differences in the dental and maxillofacial structures due to human variation, development and pathology, and the alteration caused by dental treatment, which can be visually and radiographically observed. Restorative treatment, osseointegrated dental implants, fixed orthodontic and prosthetic appliances, along with dental and maxillofacial anatomy are the most informative features for the postmortem vs. antemortem comparison. The process of comparison consists of an objective identification of the consistencies and discrepancies. However, their interpretation and the final conclusion relies on the forensic odontologist knowledge and proficiency. Computer software packages such as WinID, DVI System International and UVIS can assist in the comparisons, connecting the postmortem and antemortem information and creating a ranking of possible matches. Moreover, deep machine learning models are being explored automate the comparison process. However, all comparison procedures still require the expert’s final assessment.
Open Access
Article
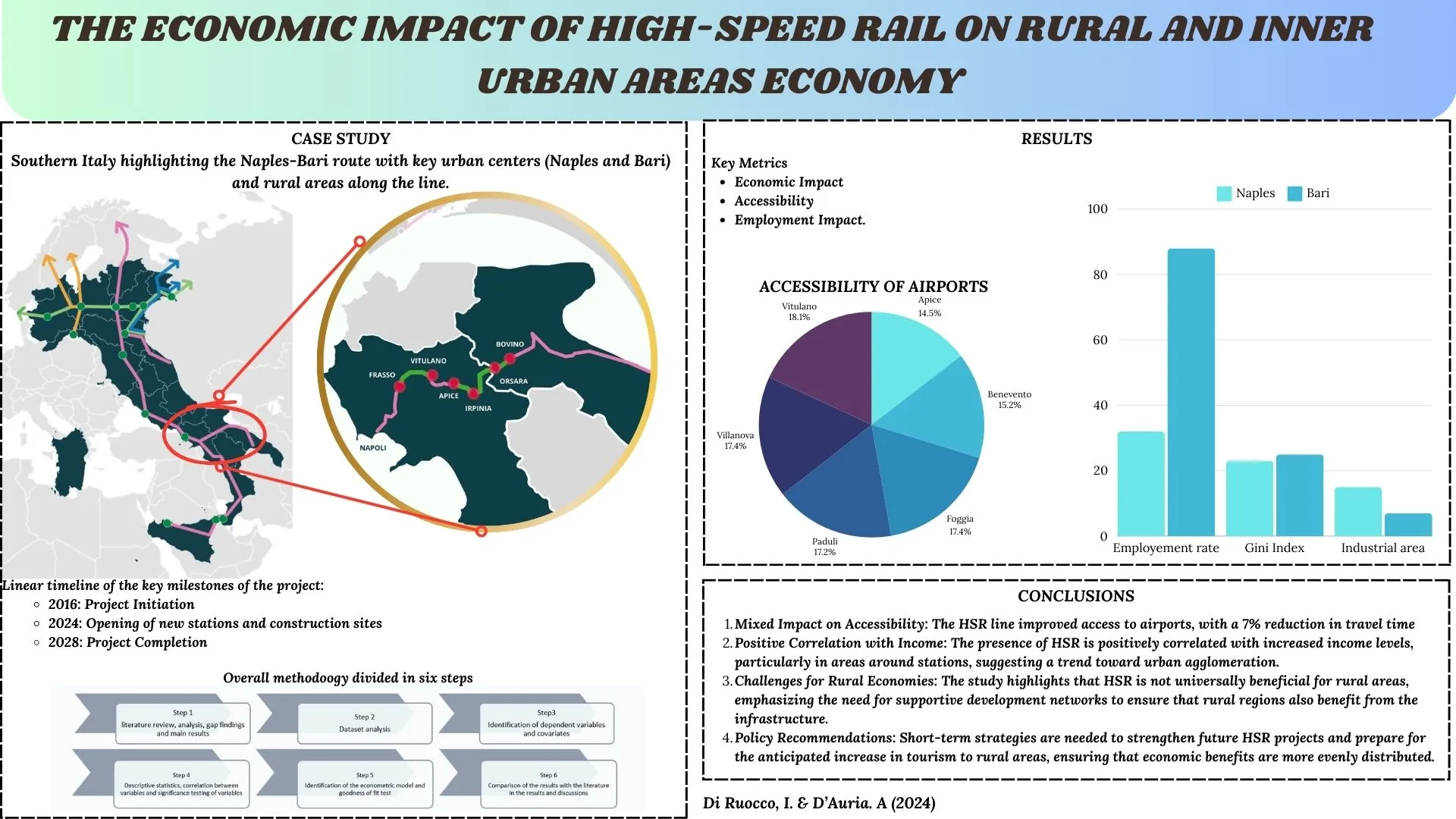
03 September 2024The Economic Impact of High-Speed Rail on Rural and Inner Urban Areas Economy: The Case Study of HSR Naples-Bari in South of Italy
High-speed rail (HSR) has revolutionized global transportation by providing fast, reliable, and efficient city-to-city travel. While its urban benefits are well-documented, the potential advantages for rural development are often overlooked. The high-speed rail project on the Naples-Bari route in Southern Italy aims to connect the urban centers of Naples in Campania and Bari in Apulia, traversing inland and rural areas. Initiated in 2016 and planned for completion in 2028, this project is anticipated to deliver numerous benefits. The purpose of this research is to examine the largely overlooked high-speed rail (HSR) in Southern Italy from an economic and territorial perspective and to determine whether it can sustainably promote rural development in the areas along the railway line. This study examines whether the HSR line will enhance economic activities, strengthen industries, and improve spatial accessibility in rural areas. Using a 2020 dataset covering 25 municipalities along the railway line, including those with stations and construction sites projected to open by 2024, three regression models were employed to estimate potential improvements in income and employment. The findings indicate mixed results: access time to airports improves, decreasing by 7%, while access to ports does not see similar benefits. Income shows a positive correlation with HSR, increasing with population growth around stations, suggesting a trend towards urban agglomeration. However, the study underscores that HSR is not universally beneficial for rural economies and that supportive development networks are crucial. Policies should adopt short-term strategies to strengthen future HSR projects and prepare for the anticipated surge in mass tourism to rural areas.
Open Access
Review
02 September 2024A Review on the Application of Nanomaterials to Boost the Service Performances of Carbon-Containing Refractories
To meet the high-quality requirements for clean steel production and fully exploit the performance advantages of carbon-containing refractories, nanomaterial has been introduced into the matrix to develop advanced carbon-containing refractories. Nanomaterials, as critical additives, play a crucial role in developing novel refractories. The service performances of carbon-containing refractories are affected not only by their physical and chemical properties but also by their microstructure. This review provides a comprehensive overview of the latest research on oxide-carbon composite refractories containing nanomaterials, categorized by their composition: nanocarbons, nano oxides, and nano non-oxides. Incorporating nanomaterials can enhance the service performances of the refractories, optimizing phase composition and microstructure. Furthermore, future research directions in nanomaterial technology for carbon-containing refractories are discussed.

Open Access
Article
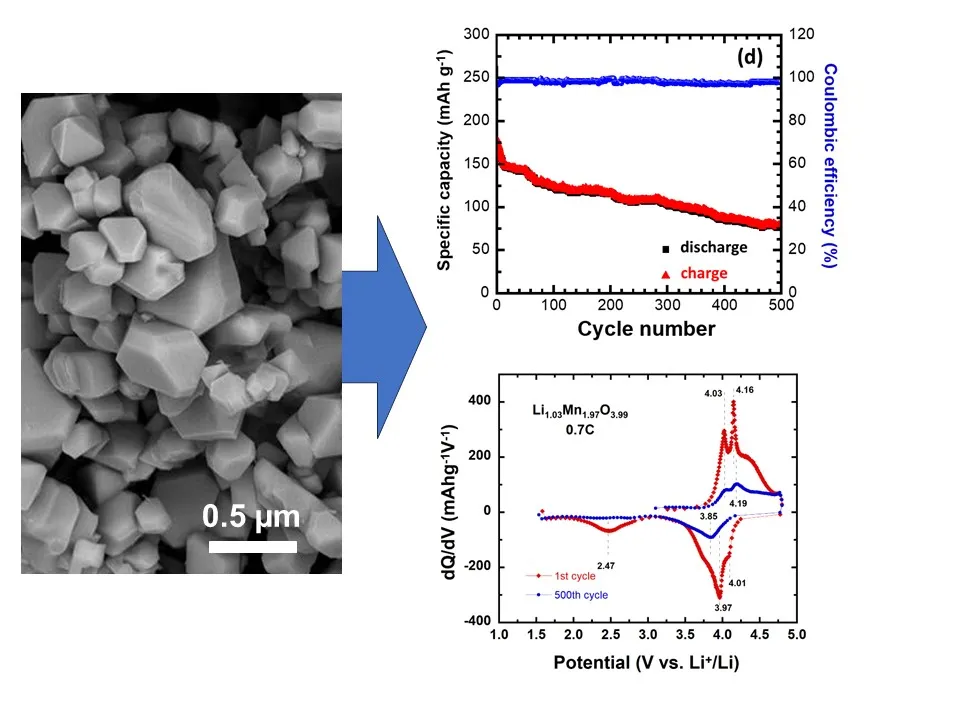
02 September 2024Lithium-Rich Spinel Cathode with Higher Energy Density for Sustainable Li-Ion Batteries Operating in Extended Potential Range
Lithium batteries pave way for rapidly reducing greenhouse gas emissions. Still there are concerns associated with battery sustainability, such as the supply of key battery materials like cobalt, nickel and carbon emissions related to their manufacture. While LiMn2O4 spinel is a common cathode material for Li-ion batteries that remove Co and Ni, studies on over-stoichiometric variants and their behavior across a broad potential range may be limited. Research in this area could provide valuable insights into the performance, stability and electrochemical characteristics of such cathodes, offering potential benefits for the development and optimization of Li-ion battery technologies. This study investigates the electrochemical behavior of Li-rich Li1+yMn2−yO4−δ (LMO, y ≈ 0.03, δ ≈ 0.01) spinel as a cathode in Li-ion batteries, focusing on the phenomenon of extra capacity under the extended operating voltage 1.5–4.8 V vs. Li+/Li. The nanostructured LMO sample synthesized by sol-gel method and calcined at 900 °C is characterized by X-ray diffraction, scanning and transmission electron microscopy and surface area measurements. The Li-rich spinel electrode delivers a specific discharge capacity of 172 mAh g−1 at 1st cycle. It retains 123 mAh g−1 at the 100th cycle (71.5% capacity retention) at current density of 100 mA g−1 current density (i.e., ~0.7 C rate). An excellent stability is obtained in the 1.5–4.8 V potential window, with a discharge capacity of 77 mAh g−1 after 500 cycles at the same current density, owing to the reduction of the Jahn-Teller effect by Li doping. These results contrast with the specific capacity of 85 mAh g−1 (1st cycle) and the capacity retention of 54.3% after 100 cycles, obtained when the cell operates in the narrow potential range of 3.0–4.5 V.
Open Access
Article
02 September 2024Risk Factors for Adverse Outcomes Following Surgical Repair of Simple Coarctation in Early Infancy: A Ten-Year Single Center Review
Coarctation of the aorta is a frequently diagnosed congenital heart defect and often requires surgical repair in early infancy. Infants born with this condition remain at risk for post-operative morbidity and reintervention within the first year of life. A single-center, retrospective chart review was performed. The protocol was approved by the Institutional Review Board (IRB-21-156, 14 August 2021). A 10-year review from January 2010 to December 2020 identified all children diagnosed with coarctation of the aorta or arch hypoplasia; without any associated major congenital cardiac pathology (i.e., simple coarctation). Reintervention-free-survival at one year for all infants who underwent surgical repair was assessed. Patient characteristics for those who did and did not experience significant adverse event (SAE) in the postoperative period were obtained and compared. A total of 105 patients diagnosed with isolated coarctation of the aorta or arch hypoplasia and who underwent surgical repair were identified. Of these, 11 patients (10%) experienced a SAE (i.e., vocal cord palsy, diaphragm palsy, chylothorax, stroke/neurological complication, need for reintubation or tracheostomy, necrotizing enterocolitis, major bleeding or thrombotic vascular complication, or reintervention) in the post-operative period. Patients who experienced a SAE were more likely to have a prenatal diagnosis of coarctation of the aorta (p = 0.03), a known genetic anomaly (p = 0.0001), or had undergone a median sternotomy approach/complex arch repair (i.e., requiring patching of the aorta) (p = 0.0001). Genetic anomaly (mainly Turner syndrome) was the only independent predictor of SAE in multivariate analysis (OR = 6.7) Follow up data at one year was available for 79 patients, with only 6 patients requiring reintervention before 1 year of age. Overall, infants who undergo surgical repair of simple coarctation of aorta have an excellent chance of reintervention-free-survival at one year post intervention. Those with a known genetic anomaly or requiring median sternotomy approach appear to have a higher risk for a SAE in the postoperative period.
Open Access
Article
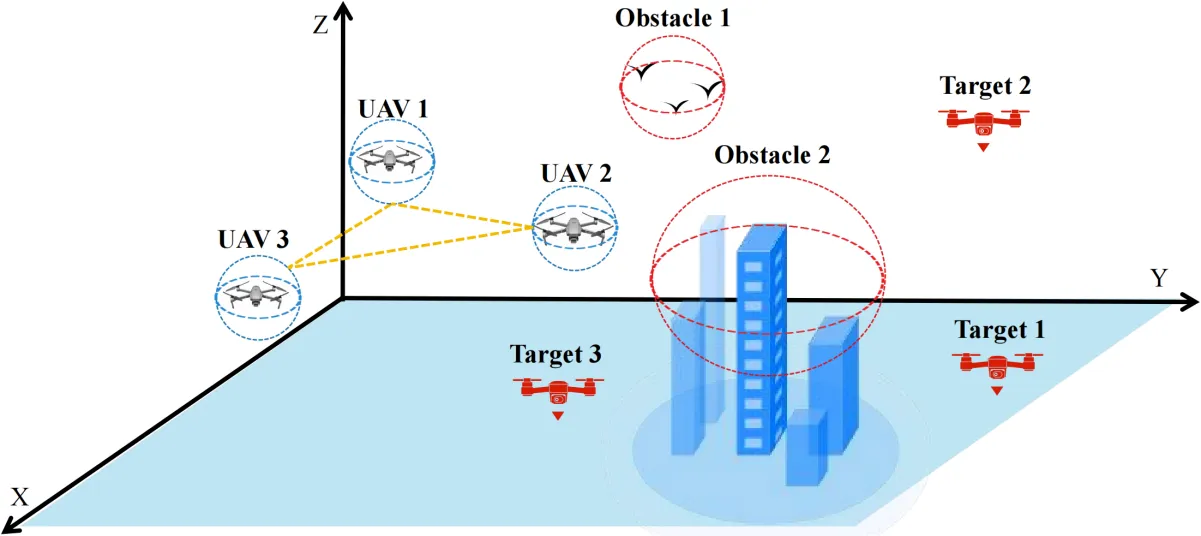
02 September 2024Multi-Robot Cooperative Target Search Based on Distributed Reinforcement Learning Method in 3D Dynamic Environments
This paper proposes a distributed reinforcement learning method for multi-robot cooperative target search based on policy gradient in 3D dynamic environments. The objective is to find all hostile drones which are considered as targets with the minimal search time while avoiding obstacles. First, the motion model for unmanned aerial vehicles and obstacles in a dynamic 3D environments is presented. Then, a reward function is designed based on environmental feedback and obstacle avoidance. A loss function and its gradient are designed based on the expected cumulative reward and its differentiation. Next, the expected cumulative reward is optimized by a reinforcement learning algorithm that makes the loss function update in the direction of the gradient. When the variance of the expected cumulative reward is lower than a specified threshold, the unmanned aerial vehicle obtains the optimal search policy. Finally, simulation results demonstrate that the proposed method effectively enables unmanned aerial vehicles to identify all targets in the dynamic 3D airspace while avoiding obstacles.
Open Access
Article
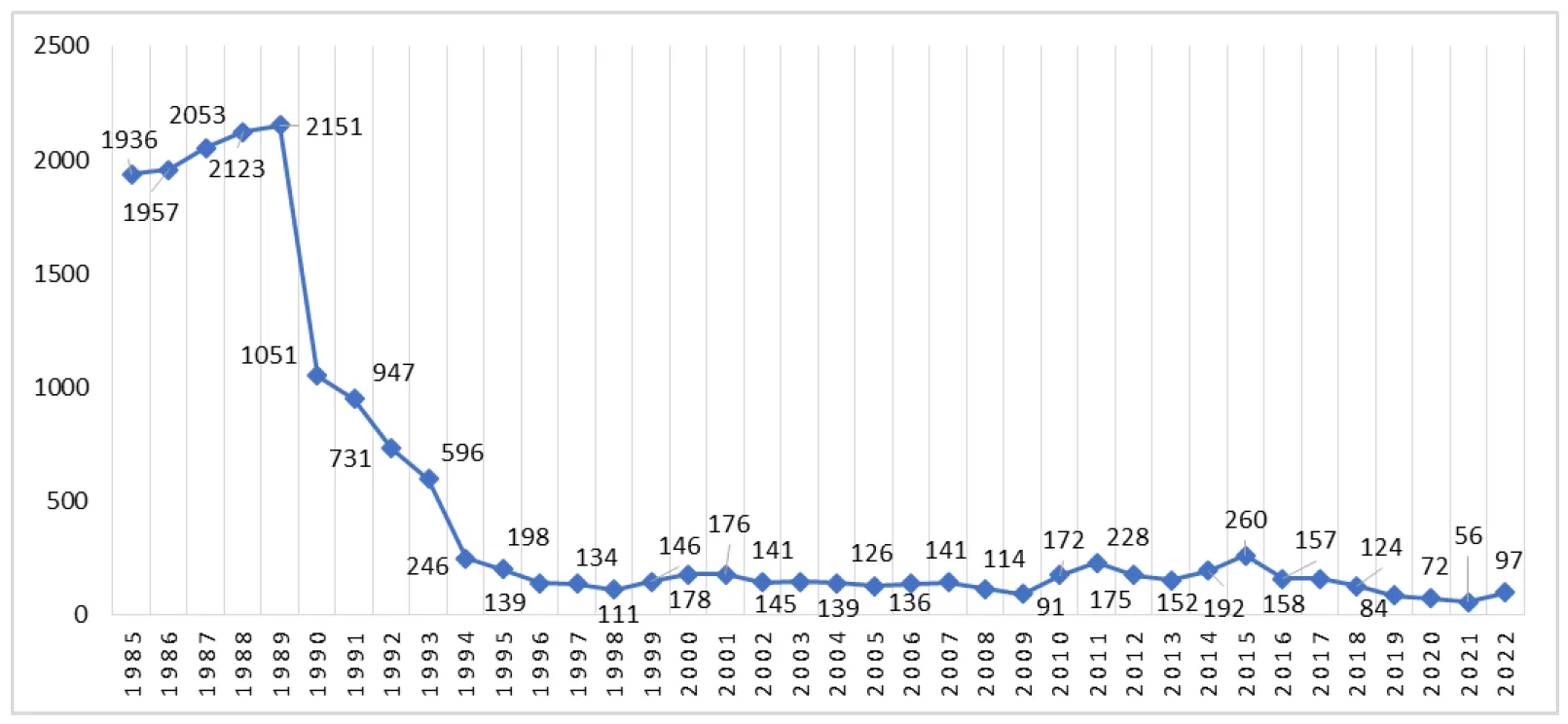
30 August 2024Model for Forecasting the Raw Material Base of the Textile Industry Based on the Analysis of the Dynamics of Production Volumes
The scientific article analyzes the dynamics of textile industry production in the USSR and the Russian Federation from 1985 to 2022 years.The article provides a fairly complete overview of modern methods of forecasting the development of objects, mainly based on time series analysis, including issues of forecasting cyclic and discontinuous processes, forecasting multidimensional objects with a correlated system of indicators. Authors calculate the forecast until 2026 year based on a bank of mathematical forecasting models implementing various monotonic nonlinear transformations both along the ordinate axis and along the abscissa axis. The criterion of the minimum variance of the forecast error was used as a criterion for selecting a specific model from the bank. The scientific value of the article lies in the fact that, for the first time, it offers a criterion for choosing a mathematical model from a set of them, which uses the minimum estimate of the variance of forecast errors for this model. This work can be considered a step towards the creation of artificial intelligence since the selection of the optimal model for a specific time series allows to obtain a training sample for it, which is fundamentally impossible to obtain without it.
Open Access
Article
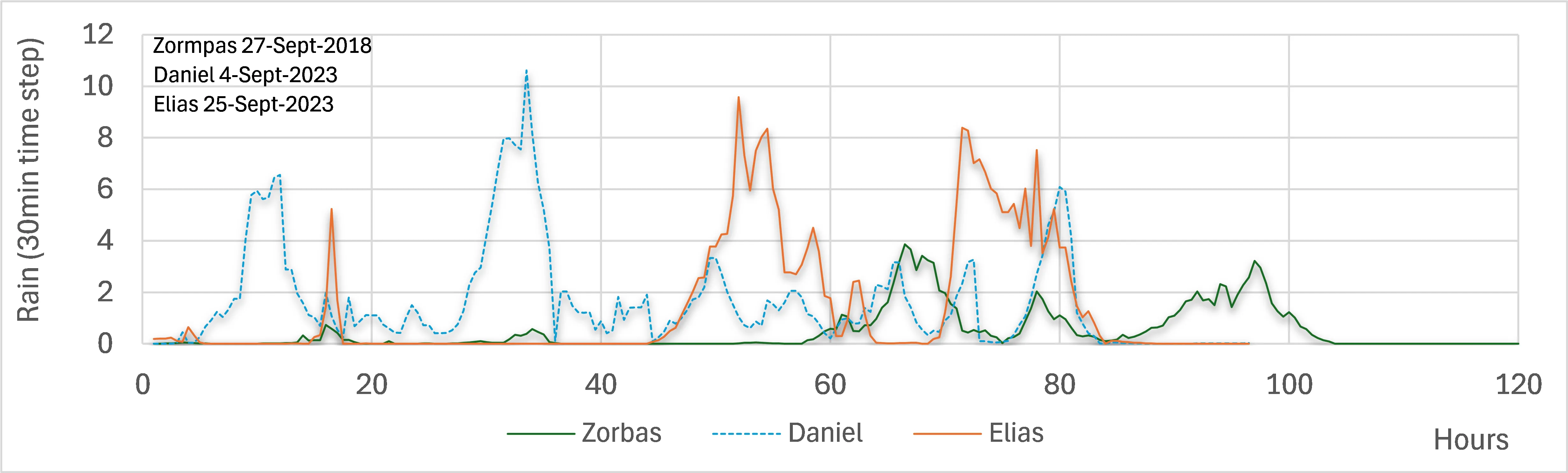
30 August 2024Fast-Track Documentation of the Alterations on the Landscape, before and after a Natural Hazard—Case Study: North Euboea Greece before and after Storms Daniel and Elias
This paper presents a methodology for fast-track documentation of landscape alterations before and after natural hazards, specifically focusing on the impacts of storms Daniel and Elias (2023) in Northern Euboea, Greece, which flooded larger areas than the storm Zorbas (2018). This happened because the plane trees had been affected by the disease Ceratocystis platani and had dried up, and the forest had burned. Therefore, the water moved faster, and in recent storms, the riverbed widened. This research aims to capture the transformed landscape rapidly by utilizing modern mapping technologies, including Google Earth, digital terrain models and drone-based photogrammetry. The methodology involves on-site inspections and the creation of three-dimensional models to document and analyze the affected areas. This approach facilitates a more comprehensive understanding of how the landscape can dynamically change due to a natural disaster. It highlights the importance of the on-site landscape inspection with sophisticated tools based on commercial equipment and open-source software.