Tools and Strategies for Engineering Bacillus methanolicus: A Versatile Thermophilic Platform for Sustainable Bioproduction from Methanol and Alternative Feedstocks
Received: 30 September 2025 Revised: 15 October 2025 Accepted: 28 October 2025 Published: 04 November 2025
© 2025 The authors. This is an open access article under the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/).
1. Introduction
The sustainable production of high-value biochemicals from methanol, a cost-effective and reduced C1 feedstock, has garnered increasing interest in the field of metabolic engineering. Among the currently explored microbial platforms, Bacillus methanolicus MGA3 (hereafter referred to as B. methanolicus) stands out as a thermophilic methylotroph that rapidly grows on methanol at 50–55 °C. At least thirteen B. methanolicus strains such as MGA3, NOA2 and PB1 have been isolated and tested for their methylotrophic growth so far [1,2,3]. However, the MGA3 strain attracted particular scientific interest thanks to its ability to overproduce l-glutamate to 59 g·L−1 during methanol fed-batch fermentations [4]. Recent advances in strain engineering have opened the possibility of utilizing alternative sustainable feedstocks, including xylose and starch. While historically recognized for its ability to overproduce amino acids, notably l-glutamate and l-lysine, recent efforts have expanded its metabolic portfolio beyond TCA cycle derivatives. In this review, we discuss recent achievements and future challenges of a B. methanolicus-based bioeconomy.
2. Developments in Tools and Technologies for Strain Engineering
Here, we outline strategies used in B. methanolicus to uncover metabolic bottlenecks and improve production strains, focusing on chemical mutagenesis, adaptive laboratory evolution (ALE), plasmid-based gene dosage, and genome editing [2,5,6,7]. Early and recent random mutagenesis studies enhanced l-lysine and l-glutamate production, and genome sequencing of mutagenized strains in combination with reverse genetics identified causal mutations in biosynthetic enzymes [2,8]. ALE has been applied to select for robust phenotypes with improved tolerance to heterologous products, revealing adaptive changes that expand production capacity [6]. Plasmid-based systems and promoter characterization enable controlled overexpression of endogenous and heterologous genes, while recent developments in CRISPR technology and suicide vectors allow manipulation of essential genes and pathways [9].
2.1. Chemical Mutagenesis and ALE as Tools to Identify Bottlenecks of Biosynthetic Pathways
In initial engineering attempts in the early 1990s, random mutagenesis approaches were used to enhance the production of native metabolites in B. methanolicus. Cells were exposed to chemical mutagens, such as ethyl methanesulfonate or N-methyl-N′-nitro-N-nitrosoguanidine, and l-lysine titers of 11·g·L−1 were obtained in methanol fed-batch fermentations from mutants resistant to the l-lysine analogue S-(2-aminoethyl)-l-cysteine [2]. The resulting mutant, B. methanolicus M168-20, displayed deregulated l-lysine feedback inhibition, and whole-genome analysis revealed mutations in the homoserine dehydrogenase hom-1 gene, which encodes a branch-point enzyme between the l-lysine and l-methionine biosynthetic pathways [8]. In recent efforts, classical random mutagenesis has been employed in combination with high-throughput screening to identify B. methanolicus clones characterized by enhanced l-glutamate production. Various mutagenic agents, including alkylating chemicals and ultraviolet irradiation, were applied. The resulting mutants exhibited ~50% higher l-glutamate titers compared to the wild type and subsequently served as chassis strains for pathway extension. Notably, the integration of recent random mutagenesis with a systems-level characterization of the mutant strain has provided a comprehensive understanding of l-glutamate biosynthesis in B. methanolicus, as described above [5].
ALE is a method in strain engineering that addresses unforeseen challenges such as energy imbalances or occurrence of toxic intermediates by selecting for robust and optimized phenotypes under defined cultivation conditions. For instance, B. methanolicus underwent ALE to enhance tolerance to the heterologous product 5-aminovalerate (5AVA), to which it is naturally susceptible. Gradual supplementation with increasing concentrations of 5AVA led to selection for mutants with alterations in iron homeostasis genes, including one encoding an iron siderophore-binding protein. Overexpression of this mutant allele in the wild-type B. methanolicus markedly improved 5AVA tolerance under high Fe2+ conditions [6]. The ALE strategy to increase 5AVA tolerance directly expanded the production capacity for methanol-based added-value chemicals in B. methanolicus. ALE was also used to increase the growth rate of the B. methanolicus strain with pBM19 plasmid integrated into the chromosome [9].
2.2. Gene Dosage Effect of Plasmid-Based Gene (Over)Expression
Plasmids are commonly used for heterologous gene expression or for the overexpression of endogenous genes. Early strain engineering efforts in B. methanolicus also relied on gene expression using the plasmid pHP13, developed as an expression vector for Bacillus subtilis [10,11]. Later, the available vector repertoire was expanded by incorporating additional replicons, including the Staphylococcus aureus plasmids pNW33N and pUB110, as well as the B. subtilis plasmid pHCMC04. Plasmids pUB110, pHP13, and pNW33N are rolling-circle replicating vectors belonging to the same incompatibility group and display approximate copy numbers of 30–50, 5–6, and ~15 per genome, respectively, in B. methanolicus. By contrast, pHCMC04 is a theta-replicating plasmid with an estimated copy number of ~3 per genome; importantly, its replication mechanism is compatible with that of rolling-circle replicating plasmids, enabling stable co-expression from two different vectors within the same host [7]. The diversification of plasmid systems for B. methanolicus represented a milestone that henceforth provided a flexible and reliable genetic toolbox, laying the foundation for metabolic engineering and multi-gene pathway optimization in this organism [12,13].
Genome-wide transcriptomic analyses in B. methanolicus have enabled the characterization of native promoter architecture and transcriptional regulation. Promoter elements were identified, including a conserved -10 motif sequence (TAtaaTaa) and a less conserved -35 consensus sequence (ttgaaa). RNA-seq further mapped genome-wide native promoter transcript abundances and transcription start sites, revealing that promoters of central metabolic genes, e.g., hps and phi, drive the highest transcript abundances, making them attractive targets for gene expression [7]. Further in vivo validation confirmed strong promoters upstream of the genes for glutamate synthase large and small subunits (gltA, gltB) and dihydroxy-acid dehydratase (ilvD) [14]. Recently, a library of synthetic promoters was constructed and characterized, leading to creation of a promoter exhibiting over double activity in comparison to commonly used mdh promoter [15]. Nevertheless, the native methanol dehydrogenase promoter (mdh) remains the standard for constitutive gene expression in B. methanolicus. Regulated gene expression has also been established, for example xylose-inducible promoters derived from Bacillus megaterium or Parageobacillus thermoglucosidasius allow controlled expression from plasmids in B. methanolicus [7,12,15]. Additionally, inducible expression can be achieved via a B. subtilis-derived pbuE riboswitch, which responds to 2-aminopurine in the culture medium [14]. Both the regulator- and riboswitch-based systems are gratuitous and titratable, providing precise control over gene expression for sensitive metabolic engineering applications. Moreover, a previously mentioned native mannitol-inducible promoter controlled by a transcriptional activator MtlR can also be used as a genetic tool, with a major limitation of mannitol being a carbon and energy source for B. methanolicus [16].
2.3. Gene Silencing and Deletion Technologies Opened Unexplored Opportunities for Strain Design and Characterization
In addition to gene overexpression, controlled repression is essential for fine-tuning metabolic pathways, reducing byproduct formation, and optimizing overall cellular performance. In this context, the Streptococcus pyogenes CRISPR-Cas9 system has emerged as a widely used tool. Gene repression is achieved via CRISPR interference (CRISPRi), in which a deactivated Cas9 protein (dCas9) binds to specific DNA sequences without generating double-stranded breaks, thereby blocking transcription initiation or elongation. This strategy is particularly advantageous for essential genes or pathways in which a genomic knockout would be deleterious, offering a level of precision and flexibility beyond traditional gene deletion methods [16]. The pNW33N plasmid was used as a basis to develop a CRISPRi system for B. methanolicus. The system has been successfully implemented to repress and functionally characterize genes involved in sporulation, H2O2 detoxification, mannitol catabolism, l-glutamate biosynthesis and the RuMP cycle in B. methanolicus [17,18]. Beyond enabling the functional characterization of the B. methanolicus physiology, CRISPRi can provide a versatile platform for rational strain development. For instance, repression of the master regulator spo0A nearly completely suppressed the sporulation phenotype, demonstrating the high efficiency of this tool [18]. This is particularly valuable as suppression of sporulation is crucial for improving performance in large-scale industrial processes, where this trait is typically undesirable. However, gene repression using CRISPRi can be genetically unstable, as it depends on dcas9 expression levels, which may fluctuate across growth phases or under varying environmental conditions. In contrast, biotechnological processes require genetically stable production hosts, making permanent genome editing a more reliable approach.
A CRISPR-Cas9 genome editing tool was developed for B. methanolicus based on the pCasPP plasmid originally designed for Paenibacillus polymyxa. The system uses a pUB110-based plasmid carrying the S. pyogenes Cas9 gene under the broad-host-range sgsE promoter from Geobacillus stearothermophilus and an sgRNA cassette driven by the constitutive gapdh promoter from Streptomyces griseus. A SpeI site enables the insertion of ~1 kb homologous arms for homology-directed repair [19]. This single-plasmid system induces Cas9-mediated double-strand breaks and leverages the native DNA repair pathways in B. methanolicus. Using this platform, the spo0A gene was deleted and replaced with a mrfp reporter gene. Moreover, deletions of the catalase-encoding katA and alanine dehydrogenase-encoding ald genes were verified by genome sequencing and corresponded to complete loss of the respective enzyme activities [20]. Furthermore, the ThermoCas9 system from Geobacillus thermodenitrificans was leveraged for better adaptation to the thermophilic nature of B. methanolicus [9]. Unlike the mesophilic S. pyogenes Cas9, ThermoCas9 exhibits broad thermal activity (20–70 °C), making it well-suited for the thermophilic physiology of B. methanolicus and enabling a robust CRISPR-based genome editing platform [9,21]. Additionally, the CRISPR/ThermoCas9 system was used to deregulate the arginine biosynthesis pathway through argR deletion, deletion of chromosomal RuMP cycle genes and integration of the pBM19 plasmid into the genome, and the physiological effects of those modifications are described in more detail in the following sections [9].
Genome modification in B. methanolicus could also be achieved using non-replicating plasmids carrying 2–6 kb homology regions to promote recombination through crossover events [15]. As proof of concept, the non-essential mutS gene was targeted using the temperature-sensitive plasmid pBMe01, which does not replicate at 60 °C, and sfGFP as a counter-selection marker [22]. In a different approach, a pUB110-derived non-replicating suicide vector was created by removing the repU gene, which encodes the plasmid replication initiation protein for Bacillus species [23]. The suicide vector carried much shorter, only ~1 kb homology regions along with selection and counterselection markers for scar-free gene deletions. With this strategy, antibiotic resistance was used for selection, while the transport of the antimetabolite 5-fluoroorotate was used for counterselection. The chromosomal upp gene, essential for uracil metabolism, was deleted using this system, producing a strain with reduced sensitivity to 5-fluorouracil, demonstrating the practical utility of the counterselection approach for robust genome engineering in B. methanolicus. Although creation of reliable genome editing tools had been sought for over two decades in B. methanolicus, the past year has witnessed remarkable progress in this field and release of novel methods for precise and efficient genome engineering in this organism (Figure 1). The historically limited availability of genome modification methods is largely due to low transformation efficiency, which has been a major bottleneck. Remarkably, all successful studies have focused on enhancing transformation efficiency, enabling the successful implementation of advanced genome editing tools in this strain.
2.4. The Next Frontier: Genome-Scale Metabolic Model-Guided Rational Strain Design
Finally, as rational strain design in bacteria increasingly relies on genome-scale metabolic models (GSMs) to guide metabolic engineering strategies, this prompted the development of two GSMs for B. methanolicus, iBmeth613 and iBM822 [5,9]. The GSM of B. methanolicus comprises 613 genes, 1020 reactions, 899 metabolites and two compartments, being cytosol and extracellular space [5]. Systems-level analyses combined with this model provide detailed insights into methanol-based production pathways, such as l-glutamate and GABA [5]. Moreover, it has been applied in flux balance analysis-guided strategies to achieve putrescine overproduction in B. methanolicus [24]. The iBM822 model comprises 822 protein-coding genes, 1730 compartmentalized reactions and 1479 metabolites, and it was used to increase l-arginine biosynthesis [9]. The recent models offer promising prospects for B. methanolicus strain engineering research.
3. Native and Non-Native Carbon Sources of B. methanolicus
We describe and discuss the native catabolic pathways in B. methanolicus, with a particular focus on methanol metabolism. Recently, its substrate range, originally limited to methanol, mannitol, glucose, and arabitol, was expanded to include xylose and starch through the introduction of heterologous pathways. While methanol utilization is what makes B. methanolicus a unique microorganism for biotechnological applications, expanding its feedstock portfolio may increase the strain’s flexibility in bioprocesses and potentially broaden its applications for co-utilization of methanol with various waste streams or other alternative feedstocks such as seaweed extracts.
3.1. Methanol Is Oxidized to Formaldehyde, Which Is Assimilated via Two Parallel Versions of the Ribulose Monophosphate Cycle
B. methanolicus is a facultative methylotroph, which can grow rapidly in media containing 0.25 g·L−1 yeast extract supplemented with 0.025 to 2 M methanol at growth rates of 0.65 ± 0.01 h−1 to 0.23 ± 0.00 h−1, respectively [25]. In methanol minimal medium, B. methanolicus grows at a rate of up to 0.46 ± 0.002 h−1. B. methanolicus assimilates methanol after its initial oxidation to formaldehyde followed by assimilation via the ribulose monophosphate (RuMP) cycle (Figure 2). Its genome consists of one chromosome (3.3 Mbp) and two plasmids, pBM19 (19 kbp) and pBM69 (69 kbp). Six of the eleven genes involved in methanol consumption have plasmid- and chromosome-borne copies. The pBM19 plasmid is essential for methylotrophic growth as its loss results in the inability to assimilate methanol [3]. Recently, pBM19 was integrated into the chromosome of B. methanolicus to prevent plasmid curing and preserve methylotrophic ability [9]. However, decreases in gene copy number and the choice of integration locus led to reduced growth rates, which were circumvented by strain development through ALE.
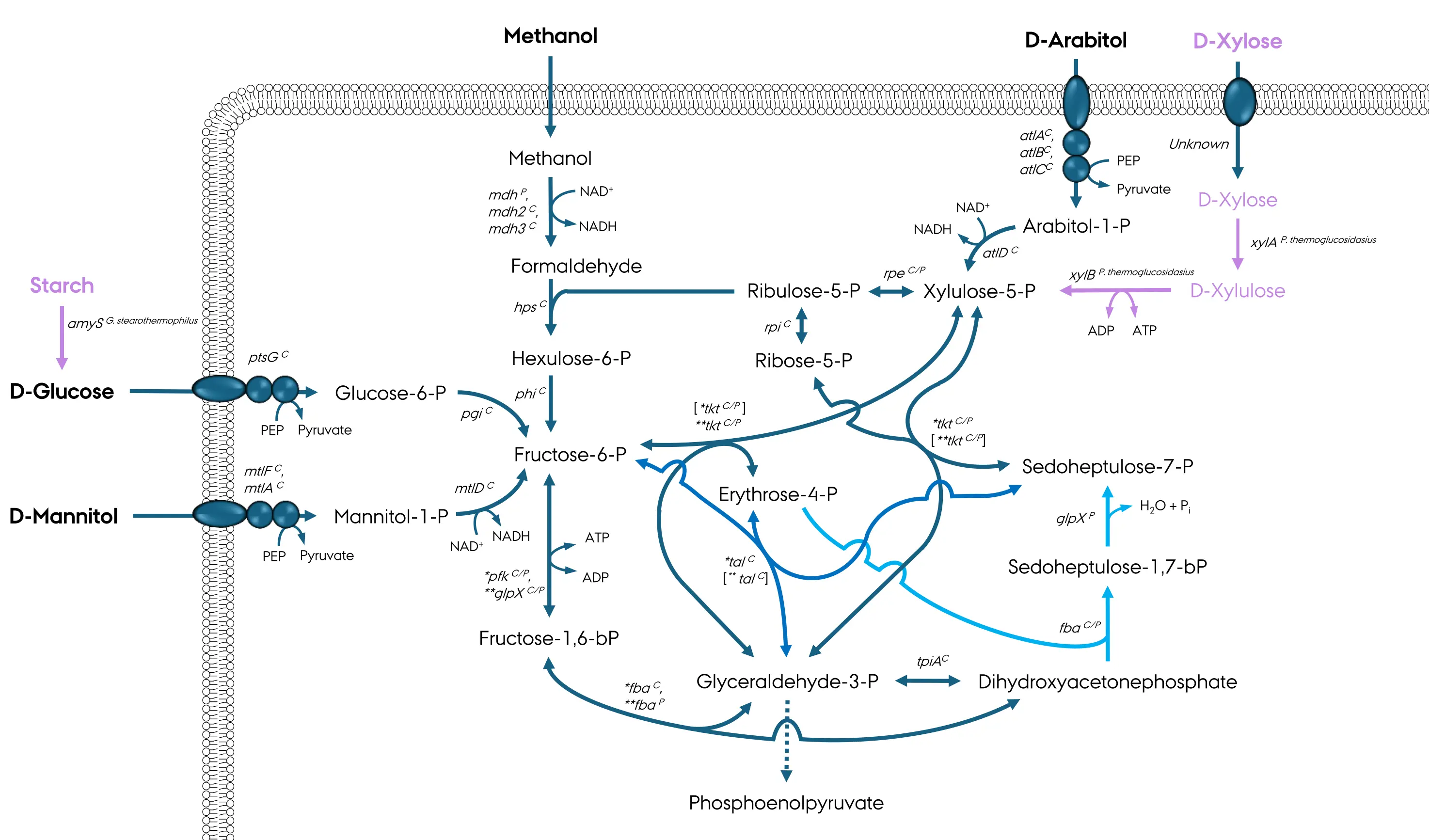
Figure 2. A schematic of uptake and assimilation of native (black font) and non-native (lilac) carbon sources in B. methanolicus. Genes: mdh, methanol dehydrogenase (EC 1.1.1.244); hps, 3-hexulose-6-phosphate synthase (EC 4.1.2.43); phi, 6-phospho-3-hexuloisomerase (EC 5.3.1.27); pfk, 6-phosphofructokinase (EC 2.7.1.11); fba, fructose-1,6-bisphosphate aldolase (EC 4.1.2.13); glpX, fructose-1,6-bisphosphatase (EC 3.1.3.1); tpiA, triosephosphate isomerase (EC 5.3.1.1); tkt, transketolase (EC 2.2.1.1); tal, transaldolase (EC 2.2.1.2); rpi, ribose-5-phosphate isomerase (EC 5.3.1.6); rpe, ribulose 5-phosphate 3-epimerase (EC 5.1.3.1); mtlF, mannitol-specific phosphotransferase enzyme IIA component (EC 2.7.1.69); mtlA, PTS system mannitol-specific enzyme IIBC component (EC 2.7.1.69); mtlD, mannitol-1-phosphate 5-dehydrogenase (EC 1.1.1.17); atlA, IIA arabitol PTS component; atlB, IIB arabitol PTS component; atlC, IIC arabitol PTS component; atlD, arabitol phosphate dehydrogenase; amyS, α-amylase (EC 3.2.1.1); ptsG, PTS-glucose-specific transporter subunit IICBA (EC 2.7.1.69); pgi, glucose-6-phosphate isomerase (EC 5.3.1.9); xylA, xylose isomerase (EC 5.3.1.5); xylB, xylulose kinase (EC 2.7.1.17). One asterisk “*”, major glycolytic FBA; Two asterisks “**”, major gluconeogenetic FBA. Superscript “C”, chromosomally encoded; superscript “P”, natural plasmid pBM19 encoded; superscript “P. thermoglucosidasius”, gene heterologously expressed and native to Parageobacillus thermoglucosidasius; superscript “G. stearothermophilus”, gene heterologously expressed and native to Geobacillus stearothermophilus. Bracket encapsulation “[…]”, putative direction. Lilac arrows, heterologous reactions; dark blue arrows, common reactions; blue arrows, transaldolase rearrangement variant reactions; light blue arrows, sedoheptulose-1,7-bisphosphatase rearrangement variant reactions. Dashed arrows indicate multiple enzymatic reactions.
To date, methanol transport over the cytoplasmic membrane has not been investigated in B. methanolicus, but it can be assumed that methanol uptake is not rate-limiting as has been shown for ethanol uptake by Zymomonas mobilis [26]. In the Gram-positive non-methylotroph Corynebacterium acetophilum, methanol diffuses passively into the cell without any additional induction [27]. The ability of non-methylotrophic bacteria to take up methanol suggests that methanol may either passively diffuse across the cell membrane or that a dedicated uptake system has been conserved for energy production by methanol oxidation. A similar mechanism might function in B. methanolicus, although the presence of methanol transporters cannot be excluded since no putative genes with this function were identified in the genome of B. methanolicus to date.
In Gram-negative methylotrophs, the methanol-oxidizing methanol dehydrogenase (Mdh) is located in the periplasmic space and requires pyrroloquinoline quinone (PQQ) as its redox co-factor [28]. Deletion of the PQQ-dependent Mdh-encoding gene in Pseudomonas sp. AM1 almost entirely halts the methanol metabolism [29]. In contrast, the Fe-NAD+-dependent Mdh of Gram-positive methylotrophic Bacillus species is located in the cytoplasm [30]. This means that methanol is oxidized by Mdh inside the cell rather than adjacent to the cell membrane, and that Mdh is most likely not associated with the transport of methanol [30].
Methanol enters the cytoplasm of B. methanolicus along its concentration gradient, which is sustained by the oxidation of methanol to formaldehyde by the Fe-NAD+-dependent Mdh [31]. Methanol dehydrogenases are key enzymes for methylotrophy, and their molecular properties and thermodynamics have been extensively reviewed elsewhere [32,33]. Methanol dehydrogenases have various cofactors, each with specific advantages and drawbacks [32]. The major merit of NAD+ as a cofactor for methanol oxidation is that it serves as a carrier of high-energy electrons. This prevents the release of excess energy in form of heat or its storage as chemical bonds in toxic H2O2. On the other hand, the Gibbs free energy change (ΔG′) for methanol oxidation at ambient temperature with NAD+ as a cofactor is positive, making the reaction thermodynamically favourable only at the elevated temperatures that match the optimal growth range of B. methanolicus. This organism possesses three genes encoding Mdh, one in the pBM19 plasmid (mdh) and two in the chromosome (mdh2 and mdh3) [31]. These enzymes have a wide substrate specificity and comparable activity in the range of 0.01–0.06 U·mg−1 with methanol, although the activator protein ACT only increases Mdh activity in vitro by 3.33–10 fold [31].
Deletions of chromosomal Mdh-encoding genes, both individually and in combination, slightly reduced growth in methanol minimal medium [9]. This suggests that the plasmid-encoded Mdh is the dominant enzyme for methanol growth, likely due to the higher copy number of the plasmid-borne gene. At the same time, it was confirmed that ACT plays a non-essential role in vivo, as the deletion of the act gene did not impact cell growth in methanol minimal medium. Similarly, the heterologous expression of act in Escherichia coli showed no effect on synthetically methylotrophic growth [9,34].
After methanol oxidation by Mdh, the resulting formaldehyde can be assimilated into biomass via the RuMP cycle. Alternatively, it is oxidized to CO2 via one of the three formaldehyde dissimilation pathways, namely, the bacillithiol- and tetrahydrofolate-dependent linear pathways as well as the dissimilatory variant of RuMP cycle (reviewed in Klein et al., 2022) [35]. Those dissimilatory pathways serve as a source of energy and prevent the accumulation of lethal levels of formaldehyde.
The RuMP cycle is divided into three phases: fixation, cleavage and rearrangement [3,36]. In the first step of the fixation phase, formaldehyde is condensed with ribulose-5-phosphate (Ru5P) to hexulose-6-phosphate (H6P) by 3-hexulose-6-phosphate synthase (Hps). The next and last step of the fixation phase is isomerization of hexulose-6-phosphate to fructose-6-phosphate (F6P) by 6-phospho-3-hexuloisomerase (Phi) (Figure 2) [3]. The B. methanolicus hps and phi genes are co-transcribed as an operon essential for methanol assimilation, with their expression upregulated during methanol-based growth [4,9,10,37]. The essentiality of hps-phi operon was confirmed by deletion of the respective genes. Noteworthy, hps and phi deletions, along with all other chromosomal deletions of RuMP cycle genes mentioned in this review, were not complemented through plasmid-based expression to exclude effects of possible off-targets. In vitro studies showed that balancing the stoichiometric ratio of Mdh to Hps-Phi or creating an Mdh-Hps-Phi fusion protein shifts the pathway equilibrium toward F6P formation, showcasing that the supply of Ru5P or the combined Hps-Phi activity might be limiting for methylotrophic growth [38,39]. In fact, the hps-phi overexpression and increased Ru5P supply both increased the growth rate of B. methanolicus on methanol [10,15].
In nature, there are two variants of the cleavage phase of the RuMP cycle, wherein F6P is either phosphorylated to fructose-1,6-bisphosphate (F1,6BP) by phosphofructokinase (pfk) or converted to 2-keto-3-deoxy-6-phosphogluconate (KDPG) in a multi-reaction process catalyzed by Entner–Doudoroff enzymes. FBP and KDPG are then cleaved by aldolases to glyceraldehyde-3-phosphate (GA3P) and either dihydroxyacetone phosphate (DHAP) or pyruvate (Figure 2). Of those two variants, B. methanolicus utilizes the fructose-1,6-bisphosphate aldolase (FBA) variant. For this ability, B. methanolicus possesses a chromosomal pfkC and a pBM19-based pfkP. Deletion of the chromosomal pfkC nearly halts methylotrophic growth of B. methanolicus [9]. The pBM19-borne pfkP has not yet been deleted, but it seems to be unable to compensate for the loss of pfkC. B. methanolicus contains chromosomally (FBAC) and pBM19-(FBAP) encoded FBA [40]. Both versions of FBA can be activated by either manganese or cobalt ions, however, based on their catalytic properties, FBAC functions as the major glycolytic FBA as it exhibits 20-fold higher catalytic efficiency toward FBP cleavage than FBAP [17,40]. The catalytic efficiency of FBAP for the aldol condensation reaction with DHAP and GA3P as substrates is about 3-fold higher than that of FBAC, thus, it can be considered the major gluconeogenic FBA [40].
Rearrangement is the last phase of the RuMP cycle, and it serves to regenerate Ru5P for the fixation step [15]. B. methanolicus is the only known bacterium which possesses genes encoding both known variants of the RuMP cycle rearrangement phase [41]. In both variants, the first step is the transfer of a glycolaldehyde group from F6P to GA3P by transketolase (TKT), forming erythrose-4-phosphate (E4P) and xylulose-5-phosphate (Xu5P) (Figure 2) [42]. Thus, TKT plays a major role in the RuMP cycle with B. methanolicus harbouring two TKT encoding genes, one in the chromosome (tktC) and the other in pBM19 (tktP) [42]. While both TKTs are dependent on bivalent cations such as Mn2+, Mg2+, and Ca2+, and exhibit similar kinetic parameters for F6P, deletion of tktC does not affect methanol metabolism [9,42]. TKTP might therefore be the major TKT for methylotrophy; however, it cannot be ruled out that both enzyme versions contribute to this function. The sedoheptulose-1,7-bisphosphatase (SBPase) variant of the regeneration phase in RuMP cycle was confirmed to be active through 13C-labelling experiments [43]. Its first step is the condensation of E4P with DHAP to sedoheptulose-1,7-bisphosphate (S1,7BP) catalyzed by an aldolase (Figure 2). Both FBA enzymes have been experimentally confirmed in vitro to be bifunctional, also acting as sedoheptulose-1,7-bisphosphate aldolase (SBA). As deletion of fbaC does not affect methanol metabolism in B. methanolicus and only pBM19-encoded FBA is upregulated in methylotrophic conditions, it can be concluded that FBAP is sufficient for methylotrophy in vivo. Interestingly, CRISPRi-mediated repression of fbaC and fbaP leads to compensatory upregulation of their paralogs, suggesting their interchangeable functionality in formaldehyde assimilation. In the next step, S1,7BP undergoes dephosphorylation to sedoheptulose-7-phosphate (S7P) catalyzed by sedoheptulose-1,7-bisphosphatase (SBPase) (Figure 2) [44]. While B. methanolicus possesses two copies of the SBPase gene glpX, chromosome- and pBM19-borne, only glpXP is upregulated during methylotrophic growth and encodes an enzyme which catalyses dephosphorylation of S1,7BP. On the other hand, both the pBM19- and chromosome-borne glpX genes encode enzymes active as fructose-1,6-bisphosphatases (FBPases) [44]. Both FBPases are activated by Mn2+, but as GlpXC has higher catalytic efficiency and a lower KM for F1,6BP than GlpXP [44], it is considered the major FBPase in B. methanolicus [44]. Moreover, methanol-based growth of B. methanolicus ΔglpXC was unaffected, indicating that GlpXC is not required for methylotrophic growth [9].
In the transaldolase (TA) rearrangement phase variant, TA catalyses the transfer of a dihydroxyacetone group from F6P to E4P, producing S7P and GA3P (Figure 2) [41]. Unlike most of the enzymes of the RuMP cycle, B. methanolicus possesses only one TA encoding gene located on the chromosome (talC). While the TA variant of rearrangement phase is consistently shown to be active in metabolic flux analyses, and the TA activity is present in B. methanolicus crude extract, talC in B. methanolicus encodes a truncated protein without TA activity in vitro leaving an unanswered question regarding genetic background of TA activity [5,41]. Thus, in B. methanolicus, both the SBPase and TA variant likely function in parallel or compensate one another in case of inactivation, which is a unique feature of this bacterium [5,41].
S7P generated in both variants of the RuMP cycle rearrangement phase serves as a substrate for the reaction catalyzed by TKT. To that extent, TKT is assumed to catalyse the transfer of a glycolaldehyde unit from S7P to GA3P, but this has not been experimentally confirmed [42]. Lastly, Ru5P can be produced from ribose-5-phosphate or Xu5P by ribose-5-phosphate isomerase (rpi) and ribulose 5-phosphate 3-epimerase, respectively (rpe) (Figure 2) [45]. RPI is encoded solely by a chromosomal gene, while RPE is encoded both chromosomally (rpeC) and on plasmid pBM19 (rpeP) [4,45]. Deletion of rpeC deemed this gene to be essential for methylotrophic growth, even though only rpeP is upregulated in such conditions [9,37]. It remains to be shown whether RPEP is required for methylotrophic growth or instead contributes by enhancing growth with methanol above the level supported by RPEC.
3.2. Rapid Growth of B. methanolicus on Mannitol Emphasizes Its Potential as a Cell Factory Using Seaweed Extracts as Feedstock
Mannitol is a 6-carbon sugar alcohol which is as a source of carbon and energy for B. methanolicus with a maximal growth rate of 0.37 ± 0.01 h−1 [46]. Mannitol is taken up from the environment into the B. methanolicus cell through the mannitol-specific phosphotransferase system (PTS) with subunits IIA and IIBC encoded by mtlF and mtlA, respectively [46]. The mannitol uptake occurs with concomitant phosphorylation to mannitol-1-phosphate with phosphoenolpyruvate (PEP) serving as a phosphate group donor, catalyzed by phosphoenolpyruvate–protein phosphotransferase and phosphocarrier protein [46]. Mannitol-1-phosphate is oxidized to F6P by NAD+-dependent mannitol-1-phosphate 5-dehydrogenase encoded by mtlD [46] (Figure 2). The organization in the genome suggests monocistronic transcription of mtlA and co-transcription of mtlRFD or mtlFD as an operon and its suboperon [47]. The mtlR gene encodes a mannitol-dependent transcriptional activator MtlR, which represses transcription of mtlRFD from its upstream promoter PmtlR. Only mannitol, but no other sugar alcohol such as arabitol, serves as an inducer [7]. Similarly to growth on methanol, deletion of pfkC completely halts mannitol-based growth [9]. In addition, deletion of fbaC and phi moderately reduced the cell growth in minimal medium with mannitol. While FBAC plays a major role in glycolytic reactions, it is unclear why deletion of phi affects growth on mannitol, however, it must be stated that the phi deletion mutant was not genetically complemented, thus, unwanted secondary off target effects may confuse conclusions about PHI function. Recent work has demonstrated that B. methanolicus can grow on the mannitol-rich extracts of brown seaweed (Saccharina latissima) [48]. B. methanolicus strain heterogously expressing dehydrosqualene synthase and dehydrosqualene desaturase produced C30 terpenoids during mannitol- and seaweed-based growth [48]. Furthermore, during shake flask cultivations on 50% v/v seaweed extract and 10 g·L−1 mannitol, recombinant B. methanolicus strains were able to secrete 287 ± 13 mg·L−1 cadaverine, a titer slightly higher compared to when cultivated on methanol [48]. Moreover, when cultivated under mannitol fed-batch fermentation this strain secreted 6.3 g·L−1 of cadaverine, underscoring the potential of B. methanolicus as a host for future seaweed-based fermentations [48].
3.3. B. methanolicus Can Utilize the Sugar Alcohol Arabitol by PTS-Mediated Uptake and Phosphorylation
The 5-carbon compound arabitol is another sugar alcohol that B. methanolicus can use as a carbon and energy source, with a maximal growth rate of 0.20 ± 0.01 h−1 [46]. Arabitol is transported into the cell via an arabitol-specific PTS with three subunits encoded by atlA, atlB, and atlC [46]. During its import via PTS, arabitol is phosphorylated by phosphoenolpyruvate–protein phosphotransferase and phosphocarrier protein to arabitol-1-phosphate with PEP serving as a phosphate group donor [46]. Arabitol-1-phosphate is then oxidized by the NAD+-dependent arabitol phosphate dehydrogenase, encoded by atlD, to Xu5P as confirmed experimentally through metabolic flux analysis (Figure 2) [43,46]. In addition, atlD from B. methanolicus was shown to restore growth of a Corynebacterium glutamicum mtlD mutant with arabitol as the sole carbon source. The atlABCD suboperon belongs to a larger operon composed of nine genes, which likely contains an additional copy of the arabitol phosphate dehydrogenase gene.
3.4. PTS-Mediated Glucose Uptake in B. methanolicus Allows for Growth with Glucose and, Upon Recombinant α-Amylase Production, Enables Starch Catabolism
B. methanolicus can also utilize glucose as a source of carbon and energy, though the maximum growth rate on glucose is lower than that on methanol and mannitol and reaches 0.16 ± 0.01 h−1 [49]. Similarly to mannitol and arabitol, glucose is transported into the cell via PTS system composed of phosphoenolpyruvate–protein phosphotransferase, phosphocarrier protein, and glucose-specific transporter subunit IICBA encoded by ptsG. Concomitant with its uptake, glucose is phosphorylated to glucose-6-phosphate, which then enters glycolysis (Figure 2) [46]. The native glucose metabolism was successfully extended to starch through recombinant α-amylase production and secretion in B. methanolicus, supporting a growth rate of 0.05 ± 0.01 h−1. The maximum OD600 reached when grown on 9 g·L−1 soluble starch is approximately. 1.1 ± 0.1 compared to 7.7 ± 0.4 on 9·g·L−1 glucose [4,49]. This indicates a potential for utilization of other sugar and sugar alcohol-based polysaccharides by B. methanolicus through heterologous production of various hydrolytic enzymes [49].
3.5. Xylose Is Established as a Non-Native Carbon Source for Ribulose-5-Phosphate Regeneration
Xylose is a 5-carbon sugar, which cannot natively be catabolized by B. methanolicus [15]. Therefore, xylose is commonly used as a gratuitous inducer for heterologous gene expression in B. methanolicus from a synthetic cloning vector pBV2xp, which contains a B. megaterium-derived xylose-inducible promoter [7,15,50]. This reveals that B. methanolicus can take xylose up from the environment, although the enzymatic repertoire responsible for this process are still unknown. Recently, xylose co-utilization with methanol was established in B. methanolicus through heterologous expression of two genes from P. thermoglucosidasius: xylA and xylB coding thermophilic xylose isomerase and xylulose kinase, respectively [15]. Xylose isomerase catalyzes the conversion of xylose to xylulose, while xylulose kinase phosphorylates xylulose to Xu5P using ATP as a phosphate group donor (Figure 2). B. methanolicus supplemented with 2.5 g·L−1 xylose achieved growth rates of 0.46–0.67 h−1 with methanol at concentrations between 5–55 g·L−1 compared to 0.27–0.51 h−1 without xylose supplementation [15]. As previously mentioned, this is likely due to a higher Ru5P supply indicating that its regeneration might be limiting for methanol-based growth [15].
4. Expanding Portfolio of Methanol-Based Value-Added Compounds
We have recently reviewed B. methanolicus as a cell factory for the production of amino acids and their derivatives from methanol (see Irla and Wendisch (2022)). Since then, our understanding of carbon fluxes during l-glutamate production and mechanism of its export has increased substantially. Additionally, the product portfolio was expanded beyond amino acids and their derivatives (Table 1).
4.1. Production of Metabolites Derived from TCA Cycle Based Amino Acids
B. methanolicus MGA3 can secrete 59 g·L−1 of l-glutamate in methanol fed-batch fermentations [4]. To better understand the carbon fluxes underlying l-glutamate production, a systems-level study was recently conducted comparing the wild-type strain and the l-glutamate overproducing strain ABBM4307 generated through chemical mutagenesis [5]. The whole-genome sequencing, transcriptomics, 13C-metabolic flux analysis, and genome-scale metabolic modelling unravelled several metabolic changes in the mutant strain. These include increased carbon flux through (1) the regeneration phase of the RuMP cycle supporting recycling of Ru5P, the formaldehyde acceptor, (2) the pyruvate carboxylase, which replenishes oxaloacetate in the TCA cycle, and (3) upper reactions of the TCA cycle from oxaloacetate to α-ketoglutarate, leading to improved precursor supply. The transcriptomic analysis indicated changes in expression levels of genes involved in various metabolic processes such as cell wall metabolism, biosynthesis of vitamins, cofactors, fatty acids, and nucleotides, electron transport chain and flagella structure and function. This study suggested that l-glutamate appears to be synthesized primarily via transamination reactions that use branched-chain amino acids rather than through the classical glutamate dehydrogenase and glutamate synthase in B. methanolicus. However, the fundamental question of the specific role of the glutamate synthase (encoded by gltAB and gltA2) or glutamate dehydrogenase (encoded by yweB) in l-glutamate biosynthesis remained unanswered. Nonetheless, knock-down and overexpression data points toward glutamate synthase playing a major role in this process as the targeted repression of the gltAB operon effectively decreased l-glutamate accumulation whereas gltAB or gltA2 overexpression increased it (Figure 3) [51,52].
A small-conductance mechanosensitive channel (MscS) has been identified as playing a role in l-glutamate export in B. methanolicus [53]. Transcriptome analyses during l-glutamate overproduction revealed that the bacterium modulates lipid metabolism, suggesting changes in membrane fluidity that facilitate channel opening [53]. This hypothesis was supported by the use of surfactants in the fermentation medium, which enhanced l-glutamate secretion [54]. In silico analysis indicated that the channel is homologous to the E. coli MscS protein. Functional studies, including targeted gene repression and overexpression, demonstrated corresponding loss- and gain-of-function effects on l-glutamate export, confirming the role of the MscS channel in this process [1]. Identification of the l-glutamate export mechanism in B. methanolicus is a significant milestone for future engineering efforts, particularly for compounds derived from l-glutamate.
l-Glutamate serves as a precursor of various non-proteinogenic and proteinogenic amino acids, for example its decarboxylation leads to the formation of γ-aminobutyric acid (GABA) (Figure 3). The increased flux towards l-glutamate production in the mutant strain ABBM4307 strain was harnessed for GABA biosynthesis resulting in a titer of 13.2 g·L−1 during fed-batch fermentation using this mutant as host (Table 1) [5]. l-Glutamate is also a precursor in an eight-step l-arginine biosynthesis pathway regulated at the transcriptional level by a repressor ArgR. Deletion of argR leads to accumulation of arginine at 0.2 g·L−1 during methanol-based growth in shake flasks (Table 1) [9]. The overexpression of argB, encoding acetylglutamate kinase, known to be the rate-limiting step in arginine biosynthesis, increased l-arginine 4-fold to 0.8 g·L−1, while overexpression of other genes of the l-arginine biosynthetic pathway did not lead to changes in its biosynthesis.
The biosynthetic capabilities of B. methanolicus were recently expanded towards production of polyamines derived from l-glutamate, i.e., putrescine and spermidine [24]. Putrescine biosynthesis at 47.5 ± 0.8 μM was achieved through heterologous expression of E. coli-derived speAB, encoding arginine decarboxylase and agmatinase, respectively (Figure 3). This titer was improved 3-fold to 137.7 ± 1.8 μM (0.012 g·L−1) upon overexpression of argCJDB encoding the endogenous ornithine biosynthesis pathway in the putrescine-producing strain (Table 1). The spermidine production was achieved with the co-expression of native speE and speH encoding spermidine synthase and S-adenosylmethionine (SAM) decarboxylase, respectively, to enable aminopropyl group transfer via decarboxylated SAM (dcSAM) (Figure 3). To enhance precursor supply for spermidine biosynthesis, E. coli-derived speAB was overexpressed in spermidine-producing strain, leading to a 2-fold increase in spermidine production to a final titer of 83.9 ± 2.7 μM (0.012 g·L−1) (Table 1). Future efforts may explore the biosynthesis of higher-chain or functionalized polyamines with applications in polymer chemistry, plant growth promotion, and biomedical materials.
Besides l-glutamate and its derivatives, l-lysine is another highly relevant target for methanol-based production, particularly considering that stoichiometrically the maximum methanol-based l-lysine production of 0.82 g·g−1 in B. methanolicus is comparable to the maximum glucose-based l-lysine production in C. glutamicum [55]. While B. methanolicus wild-type only secretes 0.4 g·L−1 l-lysine in optimized fed-batch methanol fermentations, the classically obtained mutant NOA2#13A52-8A66 overproduces l-lysine up to 65 g·L−1 [11]. Not only is l-lysine itself a product of interest, but it also serves as a precursor for various valuable compounds like 1,5-diaminopentane (cadaverine), a monomer of bioplastics. The wild-type B. methanolicus heterologously expressing E. coli-derived cadA produces cadaverine titers of 10.2 g·L−1 without l-lysine accumulation in a high-cell density fed-batch methanol fermentation [7,56]. The l-lysine biosynthesis is strongly regulated in the wild-type B. methanolicus, and accumulations of high titers of cadaverine in this genetic background strain can be explained by the metabolic pull caused by the activity of lysine decarboxylase, which deregulates feedback inhibition [56]. Furthermore, the feasibility of mannitol and seaweed as feedstocks for cadaverine production by recombinant B. methanolicus was demonstrated [48].
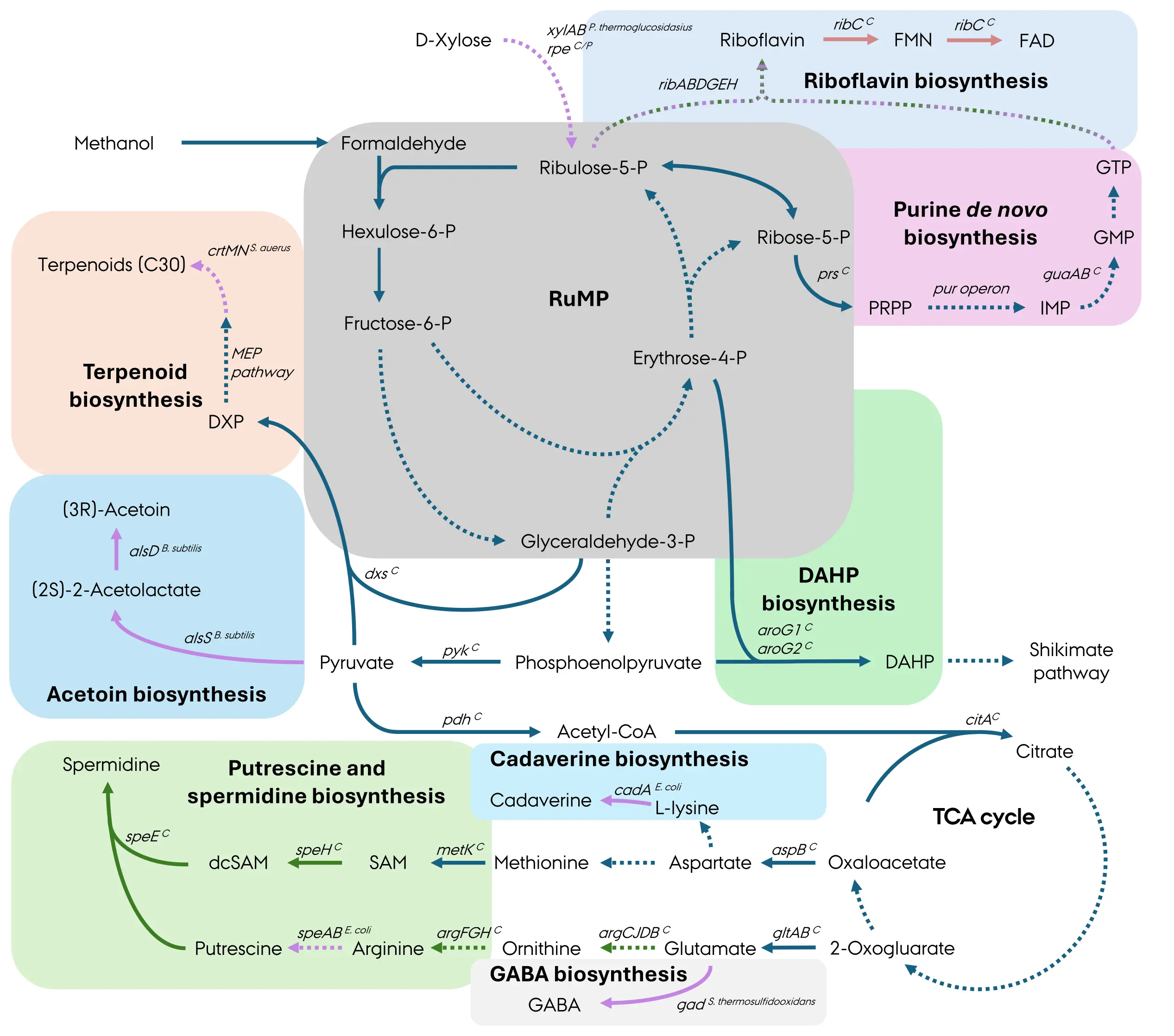
Figure 3. Biosynthesis overview of value-added compounds derived from central carbon metabolism in B. methanolicus. Pathways: RuMP cycle, ribulose monophosphate cycle; TCA, tricarboxylic acid cycle; MEP, methylerythritol-phosphate pathway. Metabolites: GTP, guanosine-5′-triphosphate; GMP, guanosine-5′-monophosphate; IMP, inosine-5′-monophosphate; PRPP, 5-phospho-α-d-ribosyl-1-pyrophosphate; DAHP, 3-deoxy-arabinoheptulosonate-7-phosphate; DXP, 1-deoxy-xylulose-5-phosphate; SAM, S-adenosylmethionine; dcSAM, decarboxylated S-adenosylmethionine. Genes: mdh, methanol dehydrogenase (EC 1.1.1.244); hps, 3-hexulose-6-phosphate synthase (EC 4.1.2.43); phi, 6-phospho-3-hexuloisomerase (EC 5.3.1.27); pfk, 6-phosphofructokinase (EC 2.7.1.11); fba, fructose-1,6-bisphosphate aldolase (EC 4.1.2.13); rpi, ribose-5-phosphate isomerase (EC 5.3.1.6); xylA, xylose isomerase (EC 5.3.1.5); xylB, xylulose kinase (EC 2.7.1.17); prs, 5-phospho-α-d-ribosyl-1-pyrophosphate synthetase (EC 2.7.6.1); pur operon, purine biosynthesis operon; guaA, guanosine 5′-monophosphate synthetase (EC 6.3.5.2); guaB, inosine 5′-monophosphate dehydrogenase (EC 1.1.1.205); ribAB, bifunctional enzyme guanosine 5′-triphosphate cyclohydrolase II/3,4-DHBP synthase (EC 4.1.99.12/3.5.4.25); ribDG, bifunctional diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino-6-(5-phosphoribosylamino)uracil reductase (EC 3.5.4.26/1.1.1.193); ribE, riboflavin synthase (EC 2.5.1.9); ribH, 6,7-dimethyl-8-(1-d-ribityl)lumazine synthase (EC 2.5.1.78); ribC, bifunctional riboflavin kinase/FMN adenylyltransferase (EC 2.7.1.26); aroG1 and aroG2, 3-deoxy-arabinoheptulosonate-7-phosphate synthase (EC 2.5.1.54); pyk, pyruvate kinase (EC 2.7.1.40); dxs, 1-deoxy-xylulose-5-phosphate synthase (EC 2.2.1.7); crtM, dehydrosqualene synthase (EC 2.5.1.96); crtN, dehydrosqualene desaturase (EC 1.3.8.-); alsS, acetolactate synthase (EC 2.2.1.6); alsD, acetolactate decarboxylase (EC 4.1.1.5); pdh, pyruvate dehydrogenase (EC 1.2.4.1); citA, citrate synthase (EC 2.3.3.16); aspB, aspartate aminotransferase (EC 2.6.1.1); gltAB, glutamate synthase (EC 1.4.1.13); argC, N-acetyl-γ-glutamyl-phosphate reductase (EC 1.2.1.38); argJ, bifunctional glutamate N-acetyltransferase (EC 2.3.1.35); argD, acetylornithine aminotransferase (EC 2.6.1.11); argB, acetylglutamate kinase (EC 2.7.2.8); argF, ornithine carbamoyltransferase (EC 2.1.3.3); argG, argininosuccinate synthase (EC 6.3.4.5); argH, argininosuccinate lyase (EC 4.3.2.1); speA, arginine decarboxylase (EC 4.1.1.19); speB, agmatinase (EC 3.5.3.11); metK, S-adenosylmethionine synthetase (EC 2.5.1.6); speH, S-adenosylmethionine decarboxylase (EC 4.1.1.50); speE, putrescine aminopropyltransferase (EC 2.5.1.16); cadA, lysine decarboxylase (EC 4.1.1.18); gad, glutamate decarboxylase (EC 4.1.1.15). Superscript “C”, chromosomally encoded; superscript “P”, natural plasmid pBM19 encoded; superscript “P. thermoglucosidasius”, gene heterologously expressed and native to Parageobacillus thermoglucosidasius; superscript “S. aureus”, gene heterologously expressed and native to Staphylococcus aureus; superscript “B. subtilis”, gene heterologously expressed and native to Bacillus subtilis; superscript “E. coli”, gene heterologously expressed and native to Escherichia coli; superscript “S. thermosulfidooxidans”, gene heterologously expressed and native to Sulfobacillus thermosulfidooxidans. Pink arrows, heterologous reactions; dark blue arrows, common reactions; green arrows, native reactions with overexpressed genes; red arrows, native reactions with gene knockdown; green-pink arrows, both native and heterologous genes have been overexpressed. Dashed arrows indicate multiple enzyme reactions.
4.2. Production of Value-Added Compounds Derived from RuMP Cycle Intermediates
Riboflavin (vitamin B2) plays a central role in redox metabolism as the precursor of the FMN and FAD cofactors and its commercial production is well established in B. subtilis [57]. Riboflavin is synthesized de novo in some microorganisms in a pathway that begins with guanosine triphosphate (GTP) and Ru5P, the latter being an intermediate of the RuMP cycle in B. methanolicus. However, methanol-based riboflavin production has only recently been realized [50]. While overexpression of native riboflavin biosynthesis rib operon increased riboflavin titers, the heterologous expression of B. subtilis and P. thermoglucosidasius-derived rib operons improved the titers up to 0.180 ± 0.002 g·L−1 and 0.202 g·L−1, respectively, in shake flask experiments (Table 1) [15,50]. Further improvement to 369 mg·L−1 was achieved by (1) increasing Ru5P supply via supplementation of xylose in xylAB-expressing strain, (2) increasing purine supply via exchange of native promoter of the purine operon to a strong synthetic promoter and (3) decreasing riboflavin conversion to FMN and FAD via genome-based point mutation that lowered activity of bifunctional riboflavin kinase/FMN adenylyl transferase RibC (Figure 3) [15]. The engineered strain expressing the xylose metabolism operon, xylAB, was cultivated in a fed-batch fermentation with minimal xylose and methanol medium, and produced riboflavin to a high titer of 2.58 g·L−1 (Table 1) [15]. Future work will reveal if increased supply of the precursor GTP will enhance riboflavin yields even more.
Table 1. Products synthetized by genetically engineered B. methanolicus strains.
|
Products |
Genetic Modifications |
Carbon Source |
Titer in Shake Flask (g·L−1) |
Titer of Fed-Batch Fermentation (g·L−1) |
Yield (g·g−1 Methanol) |
Productivity (g·L−1·h−1) |
References |
|---|---|---|---|---|---|---|---|
|
Riboflavin |
Expression of ribDGEABH from B. subtilis |
Methanol |
0.18 |
0.52 |
- |
- |
[50] |
|
Expression of xylAB and ribDEAH from P. thermoglucosidasius, mutation of genomic ribCG199D, replacement of the purine operon promoter with pH27 promoter in the genome; |
Methanol and xylose |
0.37 |
2.58 |
- |
- |
[15] |
|
|
Acetoin |
Expression of alsSD operon from B. subtilis, maeGs from Geobacillus stearothermophilus 10 and B. methanolicus-derived aceA |
Methanol |
0.42 |
- |
0.07 |
- |
[12] |
|
GABA |
Chemical mutagenesis and screening for enhanced l-glutamate production; expression of gad from S. thermosulfidooxidans; |
Methanol |
0.16 |
13.2 |
0.04 |
- |
[5] |
|
l-Arginine |
Deletion of argR and overexpression of argB |
Methanol |
0.8 |
- |
- |
- |
[9] |
|
Putrescine |
Expression of speA, speB from E. coli and argCJDB from B. methanolicus |
Methanol |
0.012 |
- |
- |
- |
[24] |
|
Spermidine |
Expression of speA, speB from E. coli and speE, speH from B. methanolicus |
Methanol |
0.012 |
- |
- |
- |
[24] |
|
Cadaverine |
Expression of cadA from E. coli |
Methanol |
0.45 |
10.2 |
- |
- |
The shikimate pathway is another metabolic route that utilizes RuMP cycle intermediates such as E4P as its precursors [58]. It serves as the biosynthetic pathway to aromatic amino acids (l-phenylalanine, l-tyrosine, l-tryptophan) and numerous secondary metabolites. The first committed step is the formation of 3-deoxy-d-arabinoheptulosonate 7-phosphate (DAHP) from E4P and PEP by DAHP synthase (aroG) (Figure 3). E4P is an intermediate of the RuMP cycle and PEP is an intermediate of the lower glycolysis, which begins with GA3P derived from the RuMP cycle. The two native DAHP synthases from B. methanolicus encoded by aroG1, aroG2 are thermostable and exhibit substrate specificity [59]. Interestingly, AroG1 is not feedback inhibited, whereas the activity of AroG2, which contains an active chorismate mutase domain, is inhibited by chorismate and prephenate. Although no production data for DAHP or downstream shikimate derivatives in B. methanolicus have yet been reported, the availability of functional native enzymes lays the groundwork for pathway extension. Since both PEP and E4P are readily derived from glycolytic and RuMP cycle reactions, it is feasible to reconstitute and amplify the entire shikimate pathway in this host, with potential challenges such as tight regulatory feedback control. Engineering efforts could involve introducing feedback-resistant variants of DAHP synthase (aroGfbr), and integrating heterologous downstream modules for l-tyrosine or l-phenylalanine biosynthesis [60].
4.3. Production of Value-Added Compounds Derived from the Precursor Molecule Pyruvate
Acetoin (3-hydroxy-2-butanone) is a valuable precursor for solvents, food flavourings, and polymer precursors derived from pyruvate. An acetoin-producing B. methanolicus strain was constructed by heterologous expression of the alsS and alsD genes from B. subtilis [12], which encode enzymes for the conversion of pyruvate to acetolactate and subsequently to acetoin (Figure 3). In minimal methanol medium, the engineered strain reached an acetoin titer of about 0.42 g·L−1, with a yield of 0.07 g·g−1 methanol (Table 1). Acetoin synthesis competes with the TCA cycle for pyruvate, which may be mitigated by fine-tuning flux through pyruvate-forming and consuming reactions, or by dynamic regulation using inducible systems to decouple growth and production phases.
Finally, terpenoids are among the most structurally diverse natural products, with wide applications as pharmaceuticals, nutraceuticals, and biofuels. In B. methanolicus, terpenoid biosynthesis occurs via the methylerythritol phosphate (MEP) pathway, which uses pyruvate and GA3P as precursors. The production of C30 terpenoids in an engineered B. methanolicus was demonstrated in a proof-of-concept study, using not only methanol but also mannitol and seaweed extracts as raw materials [48]. Though production of C30 terpenoids was only confirmed qualitatively, not quantitatively, the functionality of the endogenous MEP pathway at 50 °C suggests high potential for future engineering. Typically, key strategies to increase terpenoid production in bacteria include: (1) overexpression of rate-limiting MEP enzymes (e.g., 1-deoxy-d-xylulose-5-phosphate (DXP) synthase, DXP reductoisomerase, isopentenyl diphosphate isomerase) and (2) introduction of heterologous terpene synthases tailored to desired products (Figure 3) [61]. In addition, the engineering of NADPH and ATP supply is crucial to support the reductive steps of MEP and terpene biosynthesis [61].
5. B. methanolicus as Source and Host for Protein Production
The robustness, thermophilic lifestyle, and natural methylotrophy of B. methanolicus make its proteins attractive for synthetic biology applications, particularly for constructing methanol-utilizing production platforms in mesophilic hosts. B. methanolicus enzymes are catalytically active at elevated temperatures, which can confer certain advantages when these genes are heterologously expressed in various bacterial species.
5.1. B. methanolicus Is a Valuable Source of Novel Enzymes
Much research on B. methanolicus has focused on characterizing genes and enzymes involved in methylotrophy, more specifically in methanol oxidation and formaldehyde assimilation and dissimilation pathways. The genome of this bacterium was sequenced in 2012 [4,47], providing access to its complete collection of genes. Transcriptome [62], proteome [37], and metabolome [63] analyses have respectively identified RNAs, proteins, and metabolites with increased abundance during methylotrophic growth. The three different methanol dehydrogenases Mdh, Mdh2, and Mdh3, and the activator protein ACT, were purified and biochemically characterized [31], as well as multiple enzymes representing the RuMP pathway [17,40,41,42,44,45]. On the one hand, this research was important for understanding methylotrophy in B. methanolicus, as described in detail above. But on the other hand, these enzymes represent valuable candidates for engineering methylotrophy into non-methylotrophic hosts, typically referred to as synthetic methylotrophy.
B. methanolicus has been instrumental in engineering synthetic methylotrophy in non-methylotrophic hosts. In particular, its methanol dehydrogenase genes have been introduced into E. coli, B. subtilis, and C. glutamicum to enable methanol oxidation into formaldehyde; the first and crucial step in bacterial methylotrophy [34,64,65]. But also the genes encoding the key RuMP cycle enzymes responsible for formaldehyde assimilation, Hps and Phi, have been co-expressed in E. coli [66]. Although most bacteria possess native enzymes representing several of the RuMP pathway reactions, ribulose-phosphate 3-epimerase (rpe), ribose-5-phosphate isomerase (rpi), transketolase (tkt), and transaldolase (tal) genes have still been heterologously expressed in these hosts to facilitate regeneration of Ru5P for continuous formaldehyde assimilation needed for functional methylotrophy (reviewed by Antoniewicz) [67].
In addition to the methylotrophy genes, several additional genes from various B. methanolicus strains have been heterologously expressed and characterized in bacterial hosts. This includes the peroxygenase P450 enzyme CYP152K6 [68], a novel cold-adapted type I pullulanase for efficient amylopectin debranching hydrolysis [69], a milk clotting enzyme MCE [70], a β-fructofuranosidase for biosynthesis of lactosucrose [71], and two DAHP synthases AroG1 and AroG2 [59].
5.2. The Methylotrophic and Thermophilic Nature of B. methanolicus Makes It a Promising Host for the Production of Proteins
Although most metabolic engineering approaches using B. methanolicus have focused on enabling bioproduction of small molecules such as amino acids and their derivatives (reviewed by Pfeifenschneider et al.) [34,72], this organism has also been explored as a potential host for heterologous protein production. While still under-developed compared to traditional hosts like E. coli and B. subtilis, B. methanolicus offers certain distinct advantages for specific applications, particularly those aiming for methanol-based bioproduction. Also, the thermophilic nature of B. methanolicus enables expression of proteins that require elevated temperatures for proper folding or catalytic activity. Over years of research, the genetic toolbox of B. methanolicus has been gradually expanded. Molecular tools such as high- and low copy number replicative plasmids, strong constitutive and inducible promoters (e.g., mannitol- and xylose-inducible expression systems), and CRISPR/Cas9-based genome editing tools have been developed to support its engineering [7,49,73]. Together, these tools enable strong and controlled gene expression and the production of functional proteins at 50 °C. Examples of model and reporter proteins that have been functionally expressed in B. methanolicus are GFP, LacZ, sfGFP, and α-amylase [49,74].
The protein secretion capabilities of this bacterium are currently under investigation, with some success in secreting heterologous proteins into the culture medium, which simplifies downstream processing. Moreover, efforts are ongoing to optimize codon usage, improve translation efficiency, and expand the host range of expression elements. Challenges remain in protein yield optimization, secretion efficiency, and expanding the genetic toolbox. Methods for methanol fed-batch cultivations in bio reactors are well established for B. methanolicus [34] that will also be highly useful for scale-up bioprocess development for heterologous protein production.
6. Major Challenges in the Establishment of B. methanolicus as an Industrial Workhorse
From vitamin and bulk chemicals to the potential synthesis of complex natural products, B. methanolicus provides a robust and versatile platform for methanol-based bioproduction. Currently, one of the major limitations in the scaling-up of methanol-based bioprocesses is securing a sufficient oxygen supply for methylotrophic microorganisms. Methanol has a high energy density due to a carbon reduction degree of 6, compared to the commonly used feedstock such as glucose with a carbon reduction degree of 4. While use of methanol as a feedstock generates the surplus of reducing power for biosynthetic pathways in comparison to glucose, methanol requires at least a double oxygen supply leading to issues with large scale fermentations [75]. However, technological advancements together with the application of high throughput approaches to optimize fermentation conditions are likely to provide rapid solutions to these practical challenges [76]. Moreover, currently there is limited knowledge about transport proteins in B. methanolicus, regarding substrate uptake, export of low-molecular-weight compounds, or protein secretion, which is a limitation for the strain engineering. The thermophilic nature of B. methanolicus and accordingly, high membrane fluidity may reveal new transport systems and, consequently, new roads for metabolic engineering of transport proteins as well as membrane composition.
However, despite current constraints, the expanding genetic accessibility and demonstrated production capacity should make B. methanolicus a valuable alternative candidate for future biotechnological applications. Besides its well-established position as an effective cell factory for the production of various value-added compounds from methanol, B. methanolicus is an important source for novel genes encoding thermostable proteins useful to engineer methylotrophy into non-methylotrophic hosts and other applications, and a promising host for heterologous production of proteins especially suited for methanol-based biomanufacturing under thermophilic conditions. Interestingly, this organism can also grow well on mannitol and seaweed extracts [48], and recently was engineered toward co-utilization of the seaweed carbohydrate xylose with methanol [15], further confirming its potential for sustainable bioproduction purposes. As tools for genome editing, pathway balancing, and dynamic control have greatly matured for this organism in the past year [5,15,20,23], the landscape of possible methanol-derived products will likely expand. Moreover, the integration of ALE, and systems biology approaches will help unlock high-yield production routes for a broader range of chemicals. The future of B. methanolicus as a flagship chassis for methanol-based synthetic biology is not just feasible; it is already unfolding.
Author Contributions
Conceptualization,. L.F.B. and M.I.; Writing—Original Draft Preparation, L.F.B., M.K.F., H.Z., T.B., V.F.W., M.I.; Writing—Review & Editing, L.F.B., M.K.F., H.Z., T.B., V.F.W., M.I.; Visualization, L.F.B., M.K.F.; Funding Acquisition, L.F.B., T.B., V.F.W., M.I.
Ethics Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Funding
Luciana Fernandes Brito and Markus Klitgaard Friis were funded by the Novo Nordisk Foundation, with grants number NNF24OC0094177 and NNF23OC0086520, respectively. Haowen Zhu gratefully acknowledges support of his PhD studies in Bielefeld, Germany, by scholarship 202206880008 from the China Scholarship Council (CSC).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
-
Arfman N, Dijkhuizen L, Kirchhof G, Ludwig W, Schleifer KH, Bulygina ES, et al. Bacillus methanolicus sp. nov., a new species of thermotolerant, methanol-utilizing, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 1992, 42, 439–445. [Google Scholar]
-
Schendel FJ, Bremmon CE, Flickinger MC, Guettler M, Hanson RS. l-Lysine production at 50 degrees C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl. Environ. Microbiol. 1990, 56, 963–970. [Google Scholar]
-
Brautaset T, Jakobsen ØM, Flickinger MC, Valla S, Ellingsen TE. Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus. J. Bacteriol. 2004, 186, 1229–1238. [Google Scholar]
-
Heggeset TMB, Krog A, Balzer S, Wentzel A, Ellingsen TE, Brautaset T. Genome sequence of thermotolerant Bacillus methanolicus: Features and regulation related to methylotrophy and production of l-lysine and l-glutamate from methanol. Appl. Environ. Microbiol. 2012, 78, 5170–5181. [Google Scholar]
-
Irla M, Nærdal I, Virant D, Brautaset T, Busche T, Goranovič D, et al. Systems-level analysis provides insights on methanol-based production of l-glutamate and its decarboxylation product γ-aminobutyric acid by Bacillus methanolicus. Metab. Eng. 2025, 91, 389–404. [Google Scholar]
-
Haupka C, Brito LF, Busche T, Wibberg D, Wendisch VF. Genomic and transcriptomic investigation of the physiological response of the methylotroph Bacillus methanolicus to 5-aminovalerate. Front. Microbiol. 2021, 12, 664598. [Google Scholar]
-
Irla M, Heggeset TMB, Nærdal I, Paul L, Haugen T, Le SB, et al. Genome-based genetic tool development for Bacillus methanolicus: Theta- and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production. Front. Microbiol. 2016, 7, 1481. [Google Scholar]
-
Nærdal I, Netzer R, Irla M, Krog A, Heggeset TMB, Wendisch VF, et al. l-Lysine production by Bacillus methanolicus: Genome-based mutational analysis and l-lysine secretion engineering. J. Biotechnol. 2017, 244, 25–33. [Google Scholar]
-
Liu P, Yuan Q, Yang X, Wang Q, Chang T, Bi Y, et al. A synthetic biology toolkit for interrogating plasmid-dependent methylotrophy and enhancing methanol-based biosynthesis of Bacillus methanolicus. bioRxiv 2025. doi:10.1101/2025.05.06.652373. [Google Scholar]
-
Jakobsen ØM, Benichou A, Flickinger MC, Valla S, Ellingsen TE, Brautaset T. Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J. Bacteriol. 2006, 188, 3063–3072. [Google Scholar]
-
Brautaset T, Jakobsen ØM, Degnes KF, Netzer R, Nærdal I, Krog A, et al. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for l-lysine production from methanol at 50 °C. Appl. Microbiol. Biotechnol. 2010, 87, 951–964. [Google Scholar]
-
Drejer EB, Chan DTC, Haupka C, Wendisch VF, Brautaset T, Irla M. Methanol-based acetoin production by genetically engineered Bacillus methanolicus. Green. Chem. 2020, 22, 788–802. [Google Scholar]
-
Brito LF, Irla M, Nærdal I, Le SB, Delépine B, Heux S, et al. Evaluation of heterologous biosynthetic pathways for methanol-based 5-aminovalerate production by thermophilic Bacillus methanolicus. Front. Bioeng. Biotechnol. 2021, 9, 686319. [Google Scholar]
-
Irla M, Hakvåg S, Brautaset T. Developing a riboswitch-mediated regulatory system for metabolic flux control in thermophilic Bacillus methanolicus. Int. J. Mol. Sci. 2021, 22, 4686. [Google Scholar]
-
Li B, Yang Z, Li Z, Zhang Y, Zhang L, Wang W. Enabling genetic manipulation and robustness of Bacillus methanolicus for methanol-based bio-manufacturing. Metab. Eng. 2025, 89, 121–134. [Google Scholar]
-
Schultenkämper K, Brito LF, Wendisch VF. Impact of CRISPR interference on strain development in biotechnology. Biotechnol. Appl. Biochem. 2020, 67, 7–21. [Google Scholar]
-
Schultenkämper K, Gütle DD, López MG, Keller LB, Zhang L, Einsle O, et al. Interrogating the role of the two distinct fructose-bisphosphate aldolases of Bacillus methanolicus by site-directed mutagenesis of key amino acids and gene repression by CRISPR interference. Front. Microbiol. 2021, 12, 669220. [Google Scholar]
-
Schultenkämper K, Brito LF, López MG, Brautaset T, Wendisch VF. Establishment and application of CRISPR interference to affect sporulation, hydrogen peroxide detoxification, and mannitol catabolism in the methylotrophic thermophile Bacillus methanolicus. Appl. Microbiol. Biotechnol. 2019, 103, 5879–5889. [Google Scholar]
-
Rütering M, Cress BF, Schilling M, Rühmann B, Koffas MAG, Sieber V, et al. Tailor-made exopolysaccharides—CRISPR-Cas9 mediated genome editing in Paenibacillus polymyxa. Synth. Biol. 2017, 2, ysx007. [Google Scholar]
-
Khider MLK, Irla M, López MG, Konjetzko T, Meliawati M, Schmid J, et al. CRISPR-Cas-driven HDR-/NHEJ-genome editing in Bacillus methanolicus. Front. Microbiol. 2025, submitted. [Google Scholar]
-
Mougiakos I, Mohanraju P, Bosma EF, Vrouwe V, Finger Bou M, Naduthodi MIS, et al. Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat. Commun. 2017, 8, 1647. [Google Scholar]
-
Styles MQ, Nesbitt EA, Hoffmann TD, Queen J, Ortenzi MV, Leak DJ. The heterologous production of terpenes by the thermophile Parageobacillus thermoglucosidasius in a consolidated bioprocess using waste bread. Metab. Eng. 2021, 65, 146–155. [Google Scholar]
-
Irla M, Brito LF, Langlo J, Wohlers C, Benninghaus L, Heid C, et al. Development of a markerless tool for targeted chromosome modification in the thermophilic and methylotrophic bacterium Bacillus methanolicus. Microb. Cell Factories 2025, in revision. [Google Scholar]
-
Brito LF, Arampu A, Pérez-García F, Kaya FEA, Sayar NA, Akbulut BS, et al. Model-based engineering of Bacillus methanolicus towards de novo polyamine bioproduction from methanol. New Biotechnol. 2025, 89, 91–104. [Google Scholar]
-
Bozdag A, Komives C, Flickinger MC. Growth of Bacillus methanolicus in 2 M methanol at 50 °C: The effect of high methanol concentration on gene regulation of enzymes involved in formaldehyde detoxification by the ribulose monophosphate pathway. J. Ind. Microbiol. Biotechnol. 2015, 42, 1027–1038. [Google Scholar]
-
Schoberth SM, Chapman BE, Kuchel PW, Wittig RM, Grotendorst J, Jansen P, et al. Ethanol transport in Zymomonas mobilis measured by using in vivo nuclear magnetic resonance spin transfer. J. Bacteriol. 1996, 178, 1756–1761. [Google Scholar]
-
Murooka Y, Harada T. Active transport of alcohol in Corynebacterium acetophilum. J. Bacteriol. 1974, 118, 149–154. [Google Scholar]
-
Anthony C. The Biochemistry of Methylotrophs; Academic Press Inc.: London, UK, 1982. [Google Scholar]
-
Bellion E, Kent ME, Aud JC, Alikhan MY, Bolbot JA. Uptake of methylamine and methanol by Pseudomonas sp. strain AM1. J. Bacteriol. 1983, 154, 1168–1173. [Google Scholar]
-
Arfman N, Watling EM, Clement W, van Oosterwijk RJ, de Vries GE, Harder W, et al. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch. Microbiol. 1989, 152, 280–288. [Google Scholar]
-
Krog A, Heggeset TMB, Müller JEN, Kupper CE, Schneider O, Vorholt JA, et al. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties. PLoS ONE 2013, 8, e59188. [Google Scholar]
-
Krüsemann JL, Rainaldi V, Cotton CA, Claassens NJ, Lindner SN. The cofactor challenge in synthetic methylotrophy: Bioengineering and industrial applications. Curr. Opin. Biotechnol. 2023, 82, 102953. [Google Scholar]
-
Le TK, Lee YJ, Han GH, Yeom SJ. Methanol dehydrogenases as a key biocatalysts for synthetic methylotrophy. Front. Bioeng. Biotechnol. 2021, 9, 787791. [Google Scholar]
-
Müller JEN, Meyer F, Litsanov B, Kiefer P, Potthoff E, Heux S, et al. Engineering Escherichia coli for methanol conversion. Metab. Eng. 2015, 28, 190–201. [Google Scholar]
-
Klein VJ, Irla M, Gil López M, Brautaset T, Fernandes Brito L. Unravelling formaldehyde metabolism in bacteria: Road towards synthetic methylotrophy. Microorganisms 2022, 10, 220. [Google Scholar]
-
Murrell JC, Dalton H. Methane and Methanol Utilizers; Springer: Boston, MA, USA, 1992. [Google Scholar]
-
Müller JEN, Litsanov B, Bortfeld-Miller M, Trachsel C, Grossmann J, Brautaset T, et al. Proteomic analysis of the thermophilic methylotroph Bacillus methanolicus MGA3. Proteomics 2014, 14, 725–737. [Google Scholar]
-
Price JV, Chen L, Whitaker WB, Papoutsakis E, Chen W. Scaffoldless engineered enzyme assembly for enhanced methanol utilization. Proc. Natl. Acad. Sci. USA 2016, 113, 12691–12696. [Google Scholar]
-
Fan L, Wang Y, Tuyishime P, Gao N, Li Q, Zheng P, et al. Engineering artificial fusion proteins for enhanced methanol bioconversion. Chembiochem Eur. J. Chem. Biol. 2018, 19, 2465–2471. [Google Scholar]
-
Stolzenberger J, Lindner SN, Wendisch VF. The methylotrophic Bacillus methanolicus MGA3 possesses two distinct fructose 1,6-bisphosphate aldolases. Microbiol. Read. Engl. 2013, 159, 1770–1781. [Google Scholar]
-
Pfeifenschneider J, Markert B, Stolzenberger J, Brautaset T, Wendisch VF. Transaldolase in Bacillus methanolicus: Biochemical characterization and biological role in ribulose monophosphate cycle. BMC Microbiol. 2020, 20, 63. [Google Scholar]
-
Markert B, Stolzenberger J, Brautaset T, Wendisch VF. Characterization of two transketolases encoded on the chromosome and the plasmid pBM19 of the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. BMC Microbiol. 2014, 14, 7. [Google Scholar]
-
Delépine B, López MG, Carnicer M, Vicente CM, Wendisch VF, Heux S. Charting the metabolic landscape of the facultative methylotroph Bacillus methanolicus. Msystems 2020, 5, e00745-20. [Google Scholar]
-
Stolzenberger J, Lindner SN, Persicke M, Brautaset T, Wendisch VF. Characterization of fructose 1,6-bisphosphatase and sedoheptulose 1,7-bisphosphatase from the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. J. Bacteriol. 2013, 195, 5112–5122. [Google Scholar]
-
Balzer Le S, Heggeset TMB, Haugen T, Nærdal I, Brautaset T. 6-Phosphofructokinase and ribulose-5-phosphate 3-epimerase in methylotrophic Bacillus methanolicus ribulose monophosphate cycle. Appl. Microbiol. Biotechnol. 2017, 101, 4185–4200. [Google Scholar]
-
López MG, Irla M, Brito LF, Wendisch VF. Characterization of D-arabitol as newly discovered carbon source of Bacillus methanolicus. Front. Microbiol. 2019, 10, 1725. [Google Scholar]
-
Irla M, Neshat A, Winkler A, Albersmeier A, Heggeset TMB, Brautaset T, et al. Complete genome sequence of Bacillus methanolicus MGA3, a thermotolerant amino acid producing methylotroph. J. Biotechnol. 2014, 188, 110–111. [Google Scholar]
-
Hakvåg S, Nærdal I, Heggeset TMB, Kristiansen KA, Aasen IM, Brautaset T. Production of value-added chemicals by Bacillus methanolicus strains cultivated on mannitol and extracts of seaweed Saccharina latissima at 50 °C. Front. Microbiol. 2020, 11, 680. [Google Scholar]
-
Irla M, Drejer EB, Brautaset T, Hakvåg S. Establishment of a functional system for recombinant production of secreted proteins at 50 °C in the thermophilic Bacillus methanolicus. Microb. Cell Factories 2020, 19, 151. [Google Scholar]
-
Klein VJ, Brito LF, Perez-Garcia F, Brautaset T, Irla M. Metabolic engineering of thermophilic Bacillus methanolicus for riboflavin overproduction from methanol. Microb. Biotechnol. 2023, 16, 1011–1026. [Google Scholar]
-
Bastem GM, Sayar NA, Brito LF, Brautaset T, Virant D, Sariyar Akbulut B. Development of a novel process towards an l-malate biorefinery using methanol as feedstock. Chem. Eng. Res. Des. 2024, 212, 158–167. [Google Scholar]
-
Krog A, Heggeset TMB, Ellingsen TE, Brautaset T. Functional characterization of key enzymes involved in l-glutamate synthesis and degradation in the thermotolerant and methylotrophic bacterium Bacillus methanolicus. Appl. Environ. Microbiol. 2013, 79, 5321–5328. [Google Scholar]
-
Brito LF, Luciano D, Irla M, Virant D, Courtade G, Brautaset T. Identification of MscS as a key l-glutamate exporter in Bacillus methanolicus. Microb. Biotechnol. 2025, 18, e70252. [Google Scholar]
-
Schendel FJ, Dillingham R, Hanson RS, Sano K, Matsui K. Production of Glutamate Using Wild Type Bacillus methanolicus. U.S. Patent 6,083,728, 4 July 2000. [Google Scholar]
-
Brautaset T, Jakobsen ØM, Josefsen KD, Flickinger MC, Ellingsen TE. Bacillus methanolicus: A candidate for industrial production of amino acids from methanol at 50 °C. Appl. Microbiol. Biotechnol. 2007, 74, 22–34. [Google Scholar]
-
Nærdal I, Pfeifenschneider J, Brautaset T, Wendisch VF. Methanol-based cadaverine production by genetically engineered Bacillus methanolicus strains. Microb. Biotechnol. 2015, 8, 342–350. [Google Scholar]
-
Liu Y, Zhang Q, Qi X, Gao H, Wang M, Guan H, et al. Metabolic engineering of Bacillus subtilis for riboflavin production: A review. Microorganisms 2023, 11, 164. [Google Scholar]
-
de Boer L, Vrijbloed JW, Grobben G, Dijkhuizen L. Regulation of aromatic amino acid biosynthesis in the ribulose monophosphate cycle methylotroph Nocardia sp. 239. Arch. Microbiol. 1989, 151, 319–325. [Google Scholar]
-
Gruenberg M, Irla M, Myllek S, Draths K. Characterization of two 3-deoxy-d-arabino-heptulosonate 7-phosphate synthases from Bacillus methanolicus. Protein Expr. Purif. 2021, 188, 105972. [Google Scholar]
-
Doroshenko VG, Livshits VA, Airich LG, Shmagina IS, Savrasova EA, Ovsienko MV, et al. Metabolic engineering of Escherichia coli for the production of phenylalanine and related compounds. Appl. Biochem. Microbiol. 2015, 51, 733–750. [Google Scholar]
-
Wang C, Zada B, Wei G, Kim SW. Metabolic engineering and synthetic biology approaches driving isoprenoid production in Escherichia coli. Bioresour. Technol. 2017, 241, 430–438. [Google Scholar]
-
Irla M, Neshat A, Brautaset T, Rückert C, Kalinowski J, Wendisch VF. Transcriptome analysis of thermophilic methylotrophic Bacillus methanolicus MGA3 using RNA-sequencing provides detailed insights into its previously uncharted transcriptional landscape. BMC Genom. 2015, 16, 73. [Google Scholar]
-
Carnicer M, Vieira G, Brautaset T, Portais JC, Heux S. Quantitative metabolomics of the thermophilic methylotroph Bacillus methanolicus. Microb. Cell Factories 2016, 15, 92. [Google Scholar]
-
Gao B, Zhao N, Deng J, Gu Y, Jia S, Hou Y, et al. Constructing a methanol-dependent Bacillus subtilis by engineering the methanol metabolism. J. Biotechnol. 2022, 343, 128–137. [Google Scholar]
-
Hennig G, Haupka C, Brito LF, Rückert C, Cahoreau E, Heux S, et al. Methanol-essential growth of Corynebacterium glutamicum: Adaptive laboratory evolution overcomes limitation due to methanethiol assimilation pathway. Int. J. Mol. Sci. 2020, 21, 3617. [Google Scholar]
-
Müller JEN, Heggeset TMB, Wendisch VF, Vorholt JA, Brautaset T. Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol. Appl. Microbiol. Biotechnol. 2015, 99, 535–551. [Google Scholar]
-
Antoniewicz MR. Synthetic methylotrophy: Strategies to assimilate methanol for growth and chemicals production. Curr. Opin. Biotechnol. 2019, 59, 165–174. [Google Scholar]
-
Girvan HM, Poddar H, McLean KJ, Nelson DR, Hollywood KA, Levy CW, et al. Structural and catalytic properties of the peroxygenase P450 enzyme CYP152K6 from Bacillus methanolicus. J. Inorg. Biochem. 2018, 188, 18–28. [Google Scholar]
-
Zhang SY, Guo ZW, Wu XL, Ou XY, Zong MH, Lou WY. Recombinant expression and characterization of a novel cold-adapted type I pullulanase for efficient amylopectin hydrolysis. J. Biotechnol. 2020, 313, 39–47. [Google Scholar]
-
Li L, Zheng Z, Zhao X, Wu F, Zhang J, Yang Z. Production, purification and characterization of a milk clotting enzyme from Bacillus methanolicus LB-1. Food Sci. Biotechnol. 2019, 28, 1107–1116. [Google Scholar]
-
Liu J, Ma Y, Zhang M, Lai T, Wang Y, Yang Z. Biosynthesis of lactosucrose by a new source of β-fructofuranosidase from Bacillus methanolicus LB-1. J. Biosci. Bioeng. 2023, 135, 118–126. [Google Scholar]
-
Pfeifenschneider J, Brautaset T, Wendisch VF. Methanol as carbon substrate in the bio-economy: Metabolic engineering of aerobic methylotrophic bacteria for production of value-added chemicals. Biofuels Bioprod. Biorefin. 2017, 11, 719–731. [Google Scholar]
-
Drejer EB, Hakvåg S, Irla M, Brautaset T. Genetic tools and techniques for recombinant expression in thermophilic Bacillaceae. Microorganisms 2018, 6, 42. [Google Scholar]
-
Nilasari D, Dover N, Rech S, Komives C. Expression of recombinant green fluorescent protein in Bacillus methanolicus. Biotechnol. Prog. 2012, 28, 662–668. [Google Scholar]
-
Pluschkell SB, Flickinger MC. Dissimilation of [13C]methanol by continuous cultures of Bacillus methanolicus MGA3 at 50 °C studied by 13C NMR and isotope-ratio mass spectrometry. Microbiology 2002, 148, 3223–3233. [Google Scholar]
-
Liu WC, Gong T, Wang QH, Liang X, Chen JJ, Zhu P. Scaling-up fermentation of Pichia pastoris to demonstration-scale using new methanol-feeding strategy and increased air pressure instead of pure oxygen supplement. Sci. Rep. 2016, 6, 18439. [Google Scholar]