Found 5 results
Article
14 April 2025Efficient Extracellular Production of Phospholipase D in Escherichia coli via Genetic and Process Engineering Modification
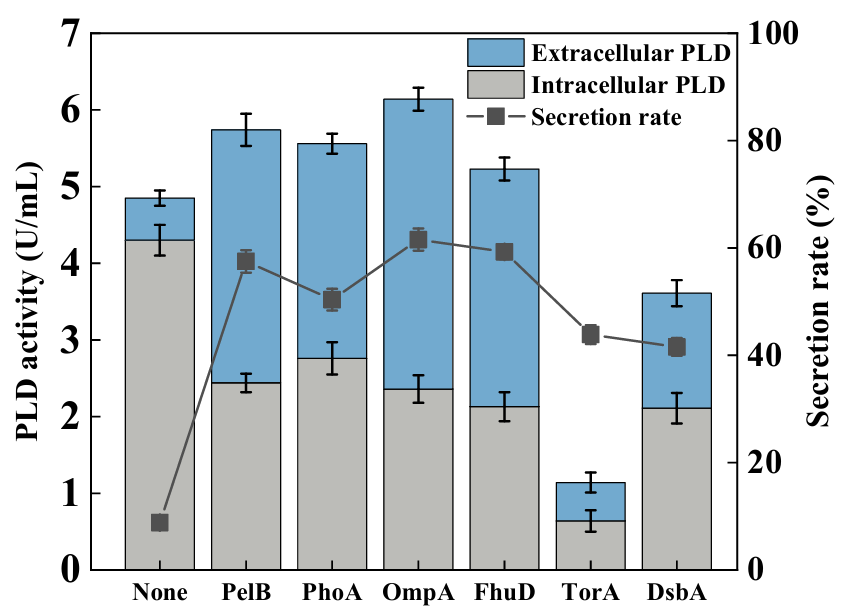
Phospholipase D (PLD) is the key enzyme in the catalytic production of rare phospholipids including phosphatidylserine. It was considered a promising method via genetic manipulation for the heterologous production of PLD in the model chassis. Few works focused on the extracellular production of PLD in engineered microbes. Herein, genetic and process engineering modification strategies were developed to achieve secretory production of PLD in Escherichia coli. The N-terminal fusion secretion signal peptide OmpA and the plasmid pBAD-gⅢC with pBAD promoter were proven to be the most effective in promoting the secretory production of PLD. Given the limitation of the cell membrane, the regulation of the key protein expression in the cell membrane as well as the addition of surfactants, were explored to accelerate the secretory production of PLD further. It was indicated that adding 0.5% (w/v) Triton X-100 was more conducive to producing PLD. Finally, fed-batch fermentation was conducted, and the maximum extracellular PLD activity achieved was 33.25 U/mL, which was the highest level reported so far. Our work demonstrated the effectiveness of genetic and process engineering strategies for the secretory production of PLD in E. coli, which provided an alternative platform for the industrial production of PLD.
Article
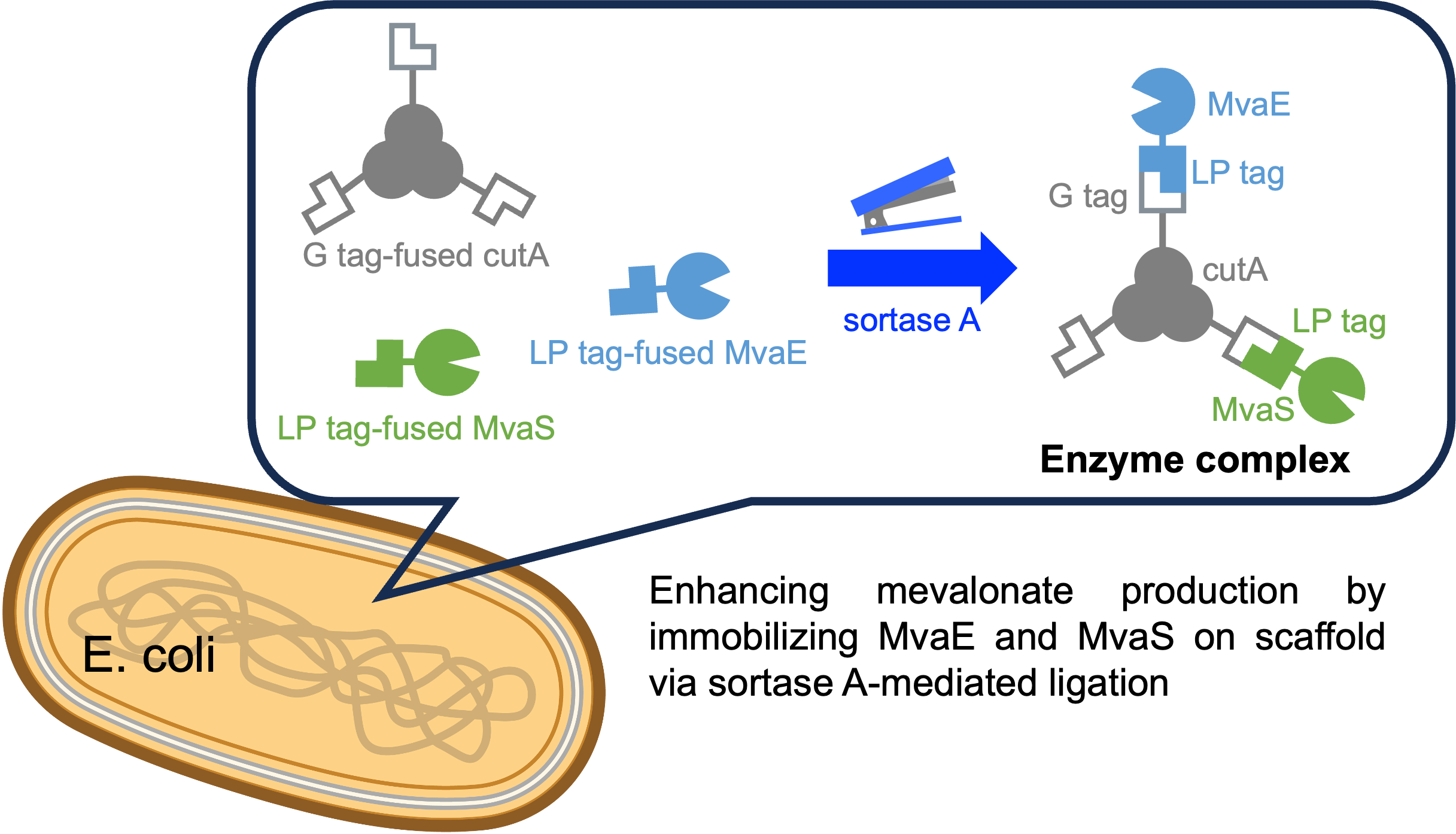
14 November 2024Sortase A-Mediated Enzyme Assembly on Multimeric Protein for Improving Mevalonate Production
Microorganisms have been extensively studied for their production of valuable chemicals. However, conventional gene fusion approaches often lack versatility and can result in enzyme inactivation. This study explored an alternative strategy for inducing metabolic channeling through sortase A-mediated ligation of metabolic enzymes. Sortase A recognizes specific amino acid sequences and selectively conjugates proteins at these sites. We focused on mevalonate production as a proof-of-concept to enhance the yield by assembling metabolic enzymes on a protein scaffold using sortase A. Although metabolic enzyme complexes were successfully formed using streptavidin as a scaffold, production did not improve. The use of CutA as a scaffold led to a 1.32-fold increase in production compared with that of the strain without the scaffold, demonstrating the efficacy of CutA in mevalonate production. These findings suggest that using sortase A to assemble metabolic enzymes onto a scaffold can effectively enhance microbial bioproduction.
Article
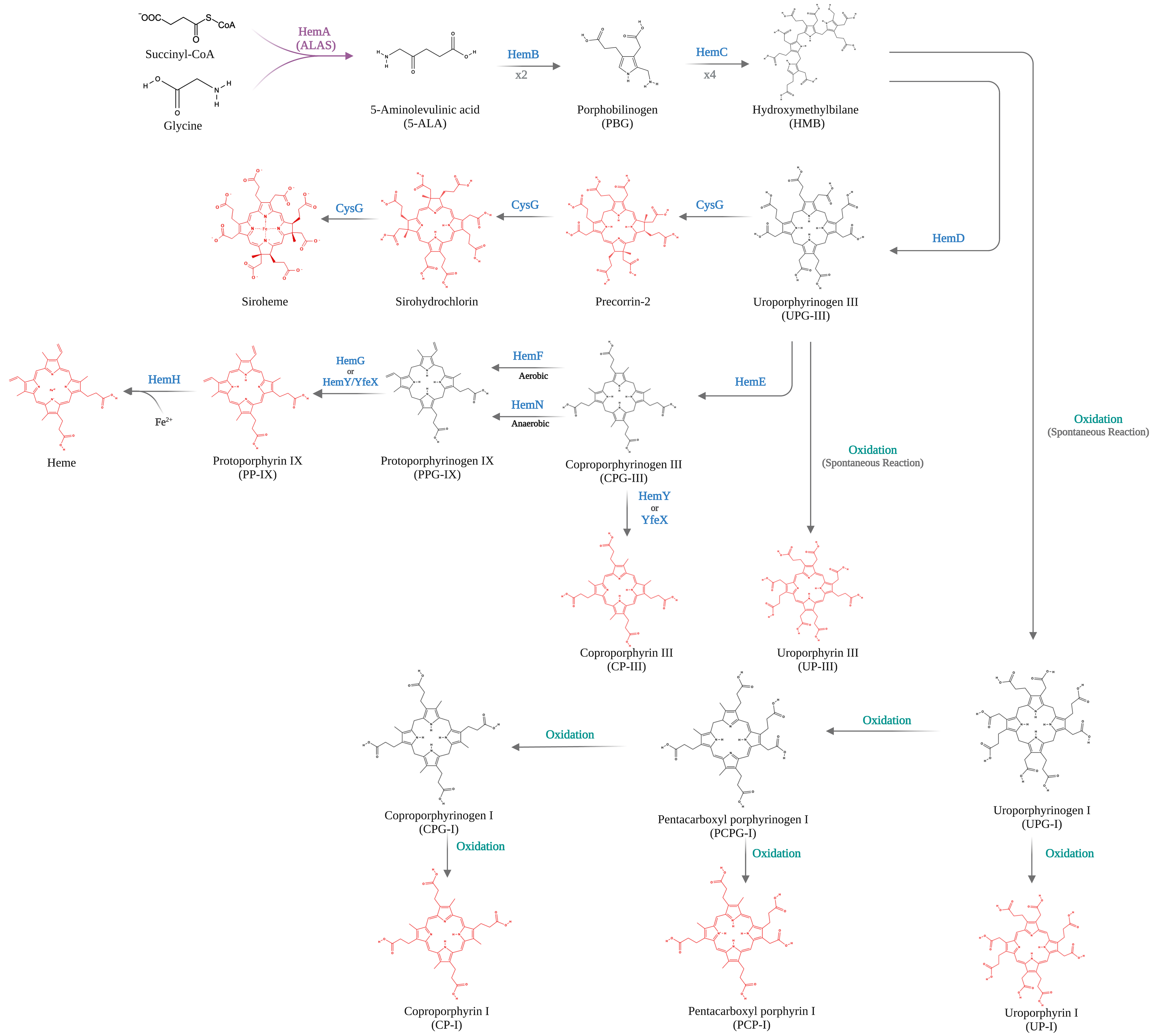
06 February 2024Bio-Based Production of Uroporphyrin in Escherichia coli
Uroporphyrin (UP) is a porphyrin compound with medical applications and a key precursor for heme biosynthesis. However, there is no biosynthetic strategy for UP production. In this study, we present a novel bioprocess for enhanced production of UP in engineered Escherichia coli. We first implemented the Shemin/C4 pathway heterologously in an E. coli strain with an enlarged intracellular pool of succinyl-CoA. Using a plasmid with the trc promoter regulating the expression of a synthesized gene operon, the effects of key pathway genes, including hemA, hemB, hemC, and hemD, on UP biosynthesis were characterized. By cultivating the resulting engineered E. coli strains in a batch bioreactor with 30 g/L glycerol under aerobic conditions, up to 901.9 mg/L UP was produced. Most of the synthesized UP was extracellularly secreted with a high purity more than 80 wt%, facilitating its downstream purification. The study paves the way for large-scale bio-based production of UP using synthetic biology and metabolic engineering strategies.
Article
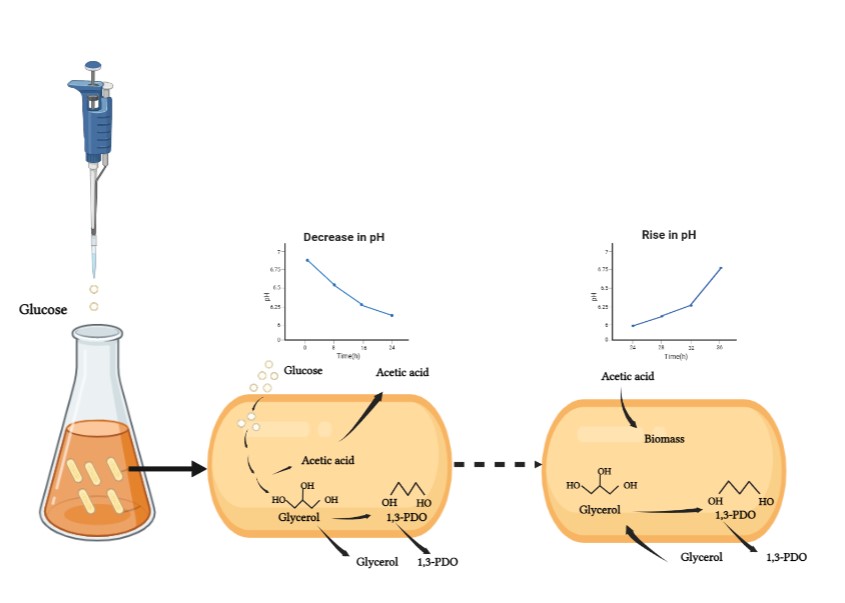
22 May 2023Fed-batch Self-regulated Fermentation of Glucose to Co-produce Glycerol and 1,3-propanediol by Recombinant Escherichia coli
As important bio-chemicals, glycerol and 1,3-propanediol (1,3-PDO) have been widely used in numerous fields, e.g., polymers, cosmetics, foods, lubricants, medicines, and so on. Bio-based 1,3-PDO is generally produced from glycerol or glucose by natural or recombinant strains, during which organic acids are always co-produced. In this work, acetic acid was also co-produced when 1,3-PDO was obtained from glucose by a recombinant strain of E. coli MG1655. Usually, a base was added to adjust the fermentation pH, resulting in the accumulation of organic salts and difficulty in the next down streaming process. Herein, a novel strategy was developed to consume the produced acetic acid by cells self in fed-batch self-regulated fermentation. The recombinant E. coli cells produced 48.33 g/L glycerol and 61.27 g/L 1,3-PDO with a total mass yield of 45.6% and without any other byproducts at the end of 5 fed-batch fermentations. The initial buffer pH, glucose concentration, pulse feeding sugar amount, time for a single batch fermentation and reducing acid were investigated by a series of comparative experiments. This fed-batch self-regulated fermentation has potential for the co-production of 1,3-PDO and glycerol, and will highlight the subsequent modification of recombinant E. coli strain by synthetic biology.
Article
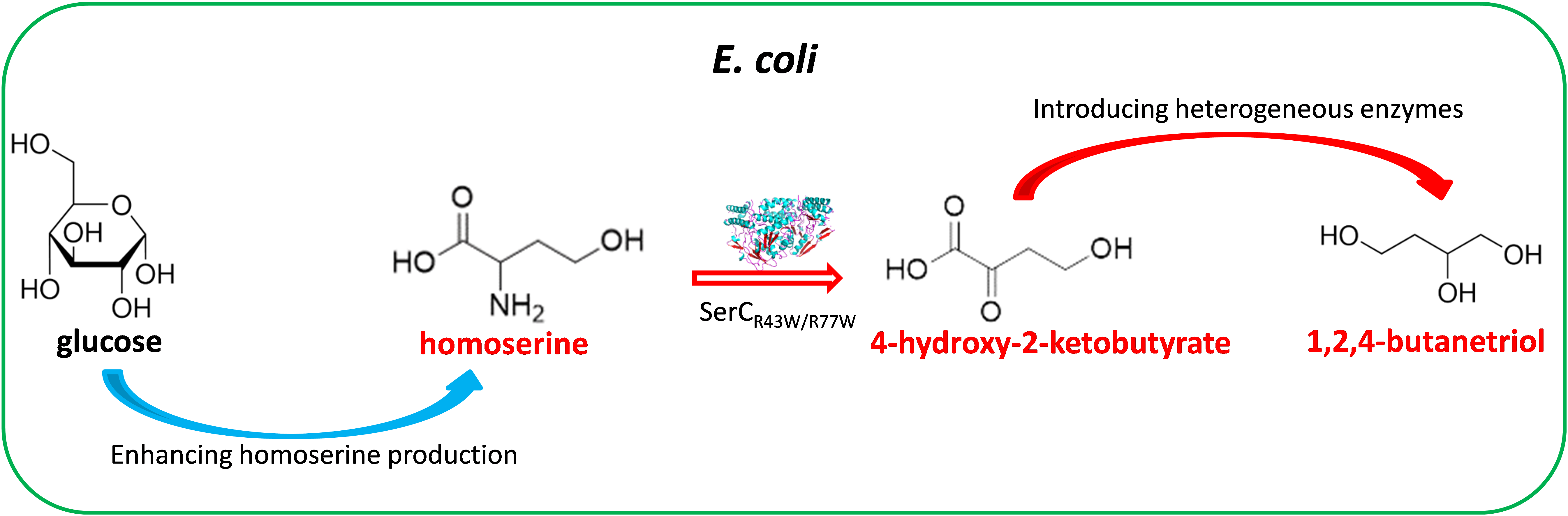
10 May 2023Development of a New 1,2,4-butanetriol Biosynthesis Pathway in an Engineered Homoserine-producing Strain of Escherichia coli
1,2,4-butanetriol (BT) is a compound of high interest with applications in pharmaceutical and materials. In this work, we designed a novel biosynthetic pathway for BT from glucose via a nonessential amino acid homoserine. This non-natural pathway used an engineered phosphoserine transaminase (SerCR42W/R77W) to achieve the deamination of homoserine to 4-hydroxy-2-oxobutanoic acid (HOBA). Three consecutive enzymes including a lactate dehydrogenase, a 4-hydroxybutyrate CoA-transferase and a bifunctional aldehyde/alcohol dehydrogenase are used to catalyze HOBA to BT. To enhance the carbon flux to homoserine, a homoserine-producing Escherichia coli was developed by improving the overexpression of two relevant key genes metL and lysC (V339A). The simultaneous overexpression of the genes encoding these enzymes for the homoserine-derived BT pathway enabled production of 19.6 mg/L BT from glucose in the homoserine-producing E. coli.