1. Introduction
Hepatic fibrosis is the result of an imbalance between production and degradation of extracellular matrix. It can be caused by various causes, such as chronic viral hepatitis infection, alcohol abuse, genetic disorders, drug intoxication, cholestasis, metabolic disorders, parasitic infections, and cryptogenic causes [
1]. The most relevant profibrogenic cells operating as cellular key drivers in the pathogenesis of liver fibrosis are hepatic stellate cells (HSCs). In the normal liver, HSC reside in the perisinusoidal space between hepatocytes and the sinusoids and exhibit a quiescent phenotype with the main function of storing vitamin A. During chronic hepatic disease, HSC progressively lose their intracellular vitamin A content, and become activated and transdifferentiate into fibrogenic myofibroblasts (MFBs) that drive the fibrogenic process by expressing large quantities of extracellular matrix compounds such as collagens. The activation process is further marked by increased expression of α-smooth muscle actin (α-SMA) and other typical markers including collagen type I (Col I), platelet-derived growth factor receptor type β (PDGFRβ), Vimentin, cysteine- and glycine-rich protein 2 (CSRP2), Fibulin, tissue inhibitor of metalloproteinase 1 (TIMP1), secreted protein acidic and rich in cysteine (SPARC), transgelin (TAGLN), and many others [
1].
Much of our current knowledge of HSC/MFB biology has been gained through primary culture studies. When HSC are cultured on the uncoated plastic, they undergo a spontaneous activation process that strongly resembles those observed in the injured liver. However, the isolation and purification of highly pure primary HSC is rather complex and the limited supply and ever-increasing demand for these cells, combined with the additional tightening of formal standards in animal welfare policy and other ethical issues, make working with these cells increasingly difficult [
2].
Moreover, challenges resulting from the demand to implement the 3R principle of “Replacement”, “Reduction”, and “Refinement” originally proposed by Russell and Burch in 1959 are additional hurdles, which oppose the use of primary cells in biomedical research [
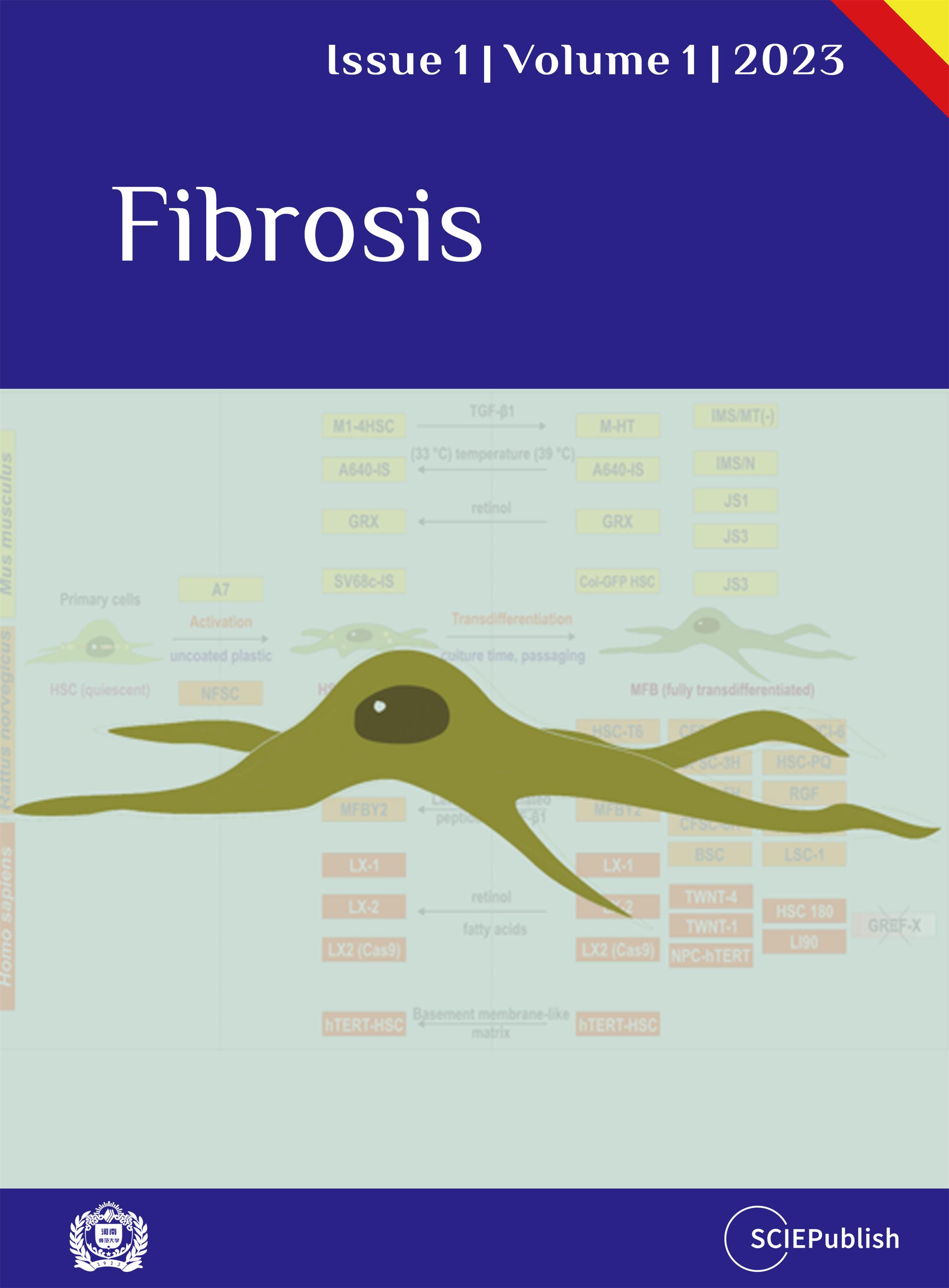
3]. Therefore, scientists have generated more than 30 infinite HSC cell lines originating from mouse, rat, or human HSC ().
. Established hepatic stellate cell lines used in biomedical research studies.
All these cell lines have specific cellular and morphological characteristics that are similar to those of primary HSC/MFB. They grow as adherent cells, express typical HSC/MFB markers, respond and transmit profibrogenic signals, and have the capacity to take up and store vitamin A. Therefore, these established HSC lines have been already important tools in investigations studying diverse aspects of HSC/MFB biology. Particularly, several of these lines have been extensively used in studies investigating aspects of profibrogenic signaling, extracellular matrix synthesis, retinoid metabolism, cellular toxicity, proliferation control, cell adhesion, and cell migration. In particular, murine cell line GRX [
10,
11], rat cell line HSC-T6 [
5,
12], different derivatives of rat cell line CFSC [
4,
13,
14], and human LX-2 cell line [
15,
16] are used in many laboratories worldwide (cf. ). The phenotype of most of these continuous growing lines determined by their overall biochemical and morphological characteristics suggests that most of these lines have to be classified as “activated HSC” or “fully transdifferentiated MFB” ().
. Classification of hepatic stellate cell lines according to their activation status. Most murine (*yellow*), rat (*orange*) and human (*red*) cell lines represent an activated or transdifferentiated phenotype. However, some of the lines (e.g., GRX, A640-IS, HSC-T6, PAV-1, MFBY2, LX-1, LX-2, hTERT-HSC) can be partly reverted into an activated phenotype by treatment with retinol, fatty acids, or the latency-associated peptide of TGF-β1. Other HSC cell lines lose some myofibroblastic features when cultured on special matrixes (e.g., hTERT-HSC) or when the temperature-sensitive large T antigen used for immortalization is inactivated by shifting the cells from the permissive to the non-permissive temperature (e.g., A640-IS). There are only a few cell lines that are not fully transdifferentiated (e.g., A7 does not proliferate under non-permissive condition; NFSC isolated from normal liver is less activated than the CFSC derivatives isolated from cirrhotic livers). Please note that the cell line GREF-X is listed in the repository of the International Cell Line Authentication Committee (ICLAC) as a misidentified cell line (registration ID: ICLAC-00123) [
9].
Although some of these cell lines can partly revert into the “activated” phenotype by treatment with high concentrations of retinol, fatty acids, latency-associated peptide, or by culturing on special basement membrane-like matrix components, none is capable to fully acquire back a quiescent cell state. In contrast to these immortalized cell lines, it has been shown that primary MFB isolated from human cirrhotic livers changed to an α-SMA negative, lipid droplet-containing quiescent morphology when re-cultured on a basement membrane-like substrate (i.e., Matrigel) [
17]. Moreover, this treatment further reduced the expression of Col I, while the expression of matrix metalloproteinase-1 (MMP-1) was induced, which underpins the reversal to an activated phenotype. Another study previously demonstrated that embedding the cells in a three-dimensionally distributed extracellular matrix prepared of rat tail type I collagen is suitable to revert their morphology, proliferation rate and functions [
18].
Each cell line has some characteristics that can be used for cell authentication. The most important one is the short tandem repeat (STR) profile that is established for some continuous growing HSC lines (e.g., GRX, Col-GFP HSC, HSC-T6, CFSC-2G, LX-2), which allows to confirm cell line identity and to identify misidentification or cross-contamination. In addition, the detection of typical HSC/MFB markers such as α-SMA, Col I, desmin, and glial fibrilliary acidic protein (GFAP) might be useful for identification. Other cell lines express the green fluorescent protein (GFP), Simian virus 40 large T-antigen (SV40T), or human telomerase reverse transcriptase (hTERT) that were stably introduced into these cells during the immortalization process. Moreover, some HSC lines have special growth properties, morphologies, or other characteristic features that are useful for cell identification.
Nowadays, several of these HSC cell lines have become in particular attractive experimental tools in studies testing compounds for their anti-fibrotic activity. Palmic acid for instance induced growth arrest and decreased α-SMA and Col I expression in the rat HSC line PAV-1 [
19]. Likewise, glycyrrhizic acid attenuated liver fibrosis in human cell line LX-2 and carbon tetrachloride (CCl4)-induced liver fibrosis, showing that results obtained in continuous growing HSC lines can provide valuable hints for the *in vivo* situation [
20]. Anti-fibrotic effects of extracellular vesicles derived from tea leaves were also reported for both HSC line LX-2 and a mouse model of CCl4-induced liver fibrosis [
21]. In the same models, a derivative of ferulic acid termed FA11 showed excellent anti-fibrotic activity as assessed by reduced HSC activation and alleviated collagen expression [
22].
Moreover, the anti-parasitic drug Ivermectin was recently shown to block activation in cultured CFSC cells and in mouse repeatedly subjected to CCl4 [
23]. In all these and many other studies, the efficacy of the drugs was majorly measured by their activity to suppress expression of genes associated with HSC activation and/or transdifferentiation (e.g., α-SMA, Col I), which underpins the key function of HSC in promoting fibrosis in response to liver injury. Unfortunately, most of the results of these preclinical studies were not translated to the clinic yet [
24].
Nevertheless, all these *in vitro* studies conducted in established HSC lines show that anti-fibrotic acting agents are majorly effective in counteracting intracellular reactive oxygen species (ROS) formation, pro-inflammatory and pro-fibrotic signaling, and extracellular matrix generation. Therefore, these studies provide the basis to develop effective drugs for treatment of human liver disease.
2. Advantages and Disadvantages for Working with Continuous HSC/MFB Lines
There are only a few HSC/MFB lines available that were spontaneously immortalized in culture without further treatment, while most of them were derived from primary HSC cultures that are either transformed with the Simian virus 40 large T-antigen (SV40T), immortalized by introducing telomerase reverse transcriptase (TERT) activity, or by exposure to intensive radiation with ultraviolet (UV) light [
6]. Consequently, respective cell lines in which genes have been artificially altered or incorporated in some way have to be classified as genetically modified cells and require a biosafety risk assessment. Nevertheless, although the primary HSC biology cannot be always accurately replicated, working with continuous growing HSC lines offers several fundamental advantages. They are cost effective, easy to handle, allow bypassing ethical concerns associated with the use of primary animal and human materials, and provide an unlimited supply of biological materials ().
. Advantages and disadvantages of working with continuous growing HSC lines versus primary cells.
Based on their clonal origin, they provide a consistent cellular source, allowing the establishment of highly reproducible results. Nevertheless, genetic drift occurring in over-passaged cultures and the immortalized phenotype *per se* could have strong impact on normal cell functions such as cell proliferation, cell cycle control, and many other native functions of HSC/MFB. In particular, the indefinite replicative capacity, escape from cell growth crisis, and potentially altered responsiveness to external and internal stimuli might provide experimental pitfalls that provoke the experimental findings other than those obtained in primary cells. Consequently, it will be of fundamental importance to reproduce and verify key experimental findings in primary cells or other suitable models. Last but not the least, it is not possible to translate findings obtained in cell lines directly to human disease. There are a multitude of factors that influence the data generated in cell culture experiments and cell lines do not fully represent what is occurring *in vivo*. Therefore, in some cases primary HSC are the better choice for more accurately reproducing cellular features of HSC in human liver disease.
3. HSC/MFB Marker Gene Expression
A common feature of the activated HSC during chronic liver disease is the expression and upregulation of typical HSC/MFB marker genes such as α-SMA, SPARC, Vimentin, and many other liver fibrosis-related core genes [
1] ().
. Immunohistochemistry of healthy and diseased human liver tissues. The tissues were stained for α-smooth muscle actin (α-SMA), secreted protein acidic and rich in cysteine (SPARC), and Vimentin representing three classical HSC/MFB markers that become increasingly expressed during hepatic insult. All images were taken from the Human Protein Atlas database [
25].
Several of these markers are positively correlated with the degree of liver fibrosis, while others such as the glial fibrillary acidic protein (GFAP) show the reduced expression in HSC during fibrosis progression. Therefore, the expression of these markers is often used to estimate the activation status of HSC/MFB or therapeutic potential of novel anti-fibrotic therapies. We have recently analyzed the transcriptomes of human cell line LX-2 (unpublished) and rat cell lines HSC-T6 [
7], CFSC-2G [
10], and PAV-1 (unpublished) by mRNA-Seq whole transcriptome analysis. Importantly, all these cell lines were shown to express typical HSC/MFB gene expression traits that include α-SMA (*ACTA2*), biglycan (*BGN*), Caldesmon 1 (*CALD1*), *COL1A1*, *COL3A3*, *SPARC*, Fibronectin (*FBN1*), Vimentin (*VIM*), and many others (). In addition, desmin representing a characteristic HSC/MFB marker in rodents was expressed in all rat HSC lines, while the expression was only rather low in human cell line LX-2. Noteworthy, several HSC/MFB markers (e.g., *PDGFRB*, Cellular communication network factor 2 (*CCN2*/*CTGF*), Retinol binding protein 1 (*RBP1*), Heart- and neural crest derivatives-expressed 2 (*HAND2*), Transgelin (*TAGLN*), and Lumican (*LUM*)) were expressed in significant different quantities in the three rat HSC lines, suggesting that each cell lines is more or less suitable for studies in which aspects of respective functions of these genes should be investigated.
For example, the highest expression of *TAGLN* was found in HSC-T6 (~764 TPM) and CFSC-2G (~655 TPM), while the mRNA content was about 39–45 fold lower in PAV-1 cells (~17 TPM). In liver, TAGLN is a typical marker of smooth muscle cells (i.e., such as HSC), correlating to the expression of *COL3A1*, FBN1, *LUM*, and *ACTA2* ( and ), suggesting that studies aiming to address functions of *TAGLN* biology should be best performed in HSC-T6 or CFSC-2G.
Contrarily, the expressions of the cellular network factors 2 (CCN
2) and 3 (CCN3) as well as PDGFRB show higher expression in PAV-1 cells compared to HSC-T6 or CFSC-2G, suggesting that gene effects of CCN
2, CCN3 or PDGFRB should be best analyzed in PAV-1 cells. This cell line was originally established in year 2002 as a new rat HSC line with capacity to convert retinol into retinoic acid [
26]. Matching to this, the expression of RBP1 is highest in cell line HSC-T6. Moreover, this is in agreement with the previous reports showing that this cell line expresses all necessary compounds such as RARα, RARβ, RARγ, RXRα, RXRβ, RXRγ, CRBP1, and ARAT, making this cell line particularly useful for studies of retinoid metabolism [
5,
12]. Certainly, these suggestions are only rough indications that need to be independently confirmed before conducting extensive studies.
. Expression of canonical HSC/MFB markers in representative immortalized human and rat HSC/MFB lines.
. Transgelin expression in liver. Single cell mRNA sequencing of various liver cell subpopulations demonstrates the highest expression of transgelin in the smooth muscle cell fraction (i.e., HSC). Data was taken from the Human Protein Atlas database [
25].
. Correlation of Transgelin (*TAGLN*) expression to other genes. Transgelin shows the highest expression in smooth muscle cells. The expression of this smooth muscle cell-specific gene correlates with the expression of Collagen type III α1 (*COL3A1*), Fibronectin 1 (*FBN1*), Lumican (*LUM*), and α-smooth muscle actin (*ACTA2*). Data was taken from the Human Protein Atlas database [
25].
4. Functional Aspects of Immortalized HSC Lines
As discussed, it is well-accepted that HSC lines are appropriate models to study general cellular and biochemical aspects of HSC biology. They express almost all relevant key markers of HSC/MFB and exhibit many features that are hallmarks of HSC, confirming that they originate from primary HSC. Nevertheless, it is obvious that all these cell lines also have pronounced differences when comparing them to primary HSC/MFB occurring in the *in vivo* situation. First, primary HSC have an overall low doubling time and can be passaged only two or three times before they enter replicative senescence and die, while established HSC lines grow significantly faster and represent unlimited self-replicating sources that can be continuously passaged. Of course, the infinite growth character of cell lines is caused by immortalized agents (e.g., SV40T, hTERT) that override the cell cycle and remove the biological brakes on proliferative control. This may *per se* modulate the general biology of the cells and affect biochemical pathways that might be relevant to HSC biology. Second, it is obvious that a clonal cell line encompasses a static phenotype, while primary HSC in culture undergo a transition from a quiescent to an activated and transdifferentiated phenotype associated with complex cellular alterations. Third, the expression repertoire of HSC lines might differ from other HSC lines and their primary counterparts. There are also some general biochemical differences between continuous growing HSC lines and primary HSC. Exemplarily, it is well-accepted that primary HSC are hardly transfectable [
27], while most of the lines are accessible for the uptake of foreign DNA. Similarly, continuous HSC lines can show divergent responsiveness towards growth factors. For instance, PDGF, representing the most effective mitogen for primary HSC [
28,
29], has only marginal stimulatory effects on proliferation on some immortal HSC lines [
8]. Moreover, single-cell RNA sequencing, cell tracing experiments, and genetic ablation experiments have shown that primary HSC form a heterogeneous cell population *in vivo* [
30,
31,
32]. It is obvious that this heterogeneity cannot be mimicked in a cell line of clonal origin. Finally, several cell lines need to be classified as the genetically modified, thereby providing potential biohazards. Therefore, the handling of those lines might require the approval from the responsible authorities and the specific institutional requirements that must be conform to legal requirements of the country in which experiments with respective cells are conducted.
Nonetheless, continuous HSC lines are key biological tools for a large abundance of studies aiming to address aspects of extracellular matrix synthesis/turnover, retinoid metabolism, fibrogenic signaling, cellular contractility/mobility, and drug efficacies. However, the above mentioned limitations in their usability and the fact that continuous cell lines are prone to genotypic and phenotypic drift limit their reasonable applicability in some studies.
5. Conclusions
Immortalized HSC/MFB lines have an unlimited life-span and stable phenotype. They are easy to handle and are widely available. Therefore, several continuous lines such as LX-2, GRX, HSC-T6 and diverse CFSC derivatives have become popular tools in biomedical research. They further allow replacing or avoiding animal use in experiments, thereby fostering the 3R principle (Replacement, Reduction and Refinement) launched over 60 years ago by Russell and Burch. Furthermore, they can be maintained in culture for an extended period of time, retaining their phenotype and function for many passages. Based on their clonal origin, continuous growing HSC/MFB cell lines provide a homogenous population, allowing the production of large quantities of uniform cells in a short period of time. Nevertheless, over-passaging, genetic drift, escape from cell growth crisis, altered responsiveness to external and internal stimuli might limit the use of these cells in some applications. Consequently, it has to be kept in mind that infinite HSC/MFB cell lines under some conditions do not behave identically equal to primary cells and can therefore not fully replace primary cells in hepatology research.
Acknowledgments
The author thanks Sabine Weiskirchen for preparing and the Graphical abstract of this Communication.
Ethics Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Funding
The laboratory of RW is supported by the German Research Foundation (grants WE2554/13-1, WE2554/15-1, WE2554/17-1) and the Interdisciplinary Centre for Clinical Research within the faculty of Medicine at the RWTH Aachen University (grant PTD 1-5). None of the funders had any role in the design of the study and decision to publish or preparation of the manuscript.
Declaration of Competing Interest
The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
1.
Acharya P, Chouhan K, Weiskirchen S, Weiskirchen R. Cellular mechanisms of liver fibrosis.
Front. Pharmacol. 2021,
12, 671640.
[Google Scholar]
2.
Meurer SK, Weiskirchen S, Tag CG, Weiskirchen R. Isolation, purification, and culture of primary murine hepatic stellate cells: An update.
Methods Mol. Biol. 2023,
2669, in press. doi:10.1007/978-1-0716-3207-9_1
[Google Scholar]
3.
Russell WMS, Burch R. The
Principles of Humane Experimental Technique; Methuen: London, UK, 1959.
4.
Nanda I, Schröder SK, Steinlein C, Haaf T, Buhl EM, Grimm DG, et al. Rat hepatic stellate cell line CFSC-2G: Genetic markers and short tandem repeat profile useful for cell line authentication.
Cells 2022,
11, 2900.
[Google Scholar]
5.
Nanda I, Steinlein C, Haaf T, Buhl EM, Grimm DG, Friedman SL, et al. Genetic characterization of rat hepatic stellate cell line HSC-T6 for in vitro cell line authentication.
Cells 2022,
11, 1783.
[Google Scholar]
6.
Friedman SL, Weiskirchen R. Working with immortalized hepatic stellate cell lines.
Methods Mol. Biol. 2023,
2669, in press. doi:10.1007/978-1-0716-3207-9_8.
[Google Scholar]
8.
González-Cuevas J, Bueno-Topete M, Armendariz-Borunda J. Urokinase plasminogen activator stimulates function of active forms of stromelysin and gelatinases (MMP-2 and MMP-9) in cirrhotic tissue.
J. Gastroenterol. Hepatol. 2006,
21, 1544–1554.
[Google Scholar]
9.
International Cell Line
Authentication Committee (ICLAC) Register of Misidentified Cell Lines. Available
online:
https://iclac.org/ (accessed on 17 March 2023).
10.
Borojevic R, Monteiro AN, Vinhas SA, Domont GB, Mourão PA, Emonard H, et al. Establishment of a continuous cell line from fibrotic schistosomal granulomas in mice livers.
In Vitro Cell Dev. Biol. 1985,
21, 382–390.
[Google Scholar]
11.
Schröder SK, Schüler HM, Petersen KV, Tesauro C, Knudsen BR, Pedersen FS, et al. Genetic and molecular characterization of the immortalized murine hepatic stellate cell line GRX.
Cells 2022,
11, 1504.
[Google Scholar]
12.
Vogel S, Piantedosi R, Frank J, Lalazar A, Rockey DC, Friedman SL, et al. An immortalized rat liver stellate cell line (HSC-T6): A new cell model for the study of retinoid metabolism in vitro.
J. Lipid Res. 2000,
41, 882–893.
[Google Scholar]
13.
Greenwel P, Schwartz M, Rosas M, Peyrol S, Grimaud JA, Rojkind M. Characterization of fat-storing cell lines derived from normal and CCl4-cirrhotic livers. Differences in the production of interleukin-6.
Lab. Invest. 1991,
65, 644–653.
[Google Scholar]
14.
Greenwel P, Rubin J, Schwartz M, Hertzberg EL, Rojkind M. Liver fat-storing cell clones obtained from a CCl4-cirrhotic rat are heterogeneous with regard to proliferation, expression of extracellular matrix components, interleukin-6, and connexin 43.
Lab. Invest. 1993,
69, 210–216.
[Google Scholar]
15.
Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis.
Gut 2005,
54, 142–151.
[Google Scholar]
16.
Weiskirchen R, Weimer J, Meurer SK, Kron A, Seipel B, Vater I, et al. Genetic characteristics of the human hepatic stellate cell line LX-2.
PLoS ONE 2013,
8, e75692.
[Google Scholar]
17.
Sohara N, Znoyko I, Levy MT, Trojanowska M, Reuben A. Reversal of activation of human myofibroblast-like cells by culture on a basement membrane-like substrate.
J. Hepatol. 2002,
37, 214–221.
[Google Scholar]
18.
Senoo H, Imai K, Sato M, Kojima N, Miura M, Hata R. Three-dimensional structure of extracellular matrix reversibly regulates morphology, proliferation and collagen metabolism of perisinusoidal stellate cells (vitamin A-storing cells).
Cell Biol. Int. 1996,
20, 501–512.
[Google Scholar]
19.
Abergel A, Sapin V, Dif N, Chassard C, Darcha C, Marcand-Sauvant J, et al. Growth arrest and decrease of alpha-SMA and type I collagen expression by palmitic acid in the rat hepatic stellate cell line PAV-1.
Dig. Dis. Sci. 2006,
51, 986–995.
[Google Scholar]
20.
Guo M, Wang Z, Dai J, Fan H, Yuan N, Gao L, et al. Glycyrrhizic acid alleviates liver fibrosis in vitro and in vivo via activating CUGBP1-mediated IFN-γ/STAT1/Smad7 pathway.
Phytomedicine 2022,
112, 154587.
[Google Scholar]
21.
Gong Q, Zeng Z, Jiang T, Bai X, Pu C, Hao Y, et al. Anti-fibrotic effect of extracellular vesicles derived from tea leaves in hepatic stellate cells and liver fibrosis mice.
Front. Nutr. 2022,
9, 1009139.
[Google Scholar]
22.
Xue T, Yue L, Zhu G, Tan Z, Liu H, Gan C, et al. An oral phenylacrylic acid derivative suppressed hepatic stellate cell activation and ameliorated liver fibrosis by blocking TGF-β1 signalling.
Liver Int. 2023,
43, 718–732.
[Google Scholar]
23.
Ying H, Li L, Zhao Y, Ni F. Ivermectin attenuates CCl4-induced liver fibrosis in mice by suppressing hepatic stellate cell activation.
Int. J. Mol. Sci. 2022,
23, 16043.
[Google Scholar]
24.
Weiskirchen R. Hepatoprotective and anti-fibrotic agents: It’s time to take the next step.
Front. Pharmacol. 2016,
6, 303.
[Google Scholar]
25.
The Human Protein Atlas. The Open Access Resource for Human Proteins. Available online:
https://www.proteinatlas.org/ (accessed on 17 March
2023).
26.
Sauvant P, Sapin V, Abergel A, Schmidt CK, Blanchon L, Alexandre-Gouabau MC, et al. PAV-1, a new rat hepatic stellate cell line converts retinol into retinoic acid, a process altered by ethanol.
Int. J. Biochem. Cell Biol. 2002,
34, 1017–1029.
[Google Scholar]
27.
Weiskirchen R, Kneifel J, Weiskirchen S, van de Leur E, Kunz D, Gressner AM. Comparative evaluation of gene delivery devices in primary cultures of rat hepatic stellate cells and rat myofibroblasts.
BMC Cell Biol. 2000,
1, 4.
[Google Scholar]
28.
Pinzani M, Gesualdo L, Sabbah GM, Abboud HE. Effects of platelet-derived growth factor and other polypeptide mitogens on DNA synthesis and growth of cultured rat liver fat-storing cells.
J. Clin. Invest. 1989,
84, 1786–1793.
[Google Scholar]
29.
Borkham-Kamphorst E, van Roeyen CR, Ostendorf T, Floege J, Gressner AM, Weiskirchen R. Pro-fibrogenic potential of PDGF-D in liver fibrosis.
J. Hepatol. 2007,
46, 1064–1074.
[Google Scholar]
30.
Magness ST, Bataller R, Yang L, Brenner DA. A dual reporter gene transgenic mouse demonstrates heterogeneity in hepatic fibrogenic cell populations.
Hepatology 2004,
40, 1151–1159.
[Google Scholar]
31.
Filliol A, Saito Y, Nair A, Dapito DH, Yu LX, Ravichandra A, et al. Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis.
Nature 2022,
610, 356–365.
[Google Scholar]
32.
Krenkel O, Hundertmark J, Ritz TP, Weiskirchen R, Tacke F. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis.
Cells 2019,
8, 503.
[Google Scholar]